Altered Nasal Microbiota Composition Associated with Development of Polyserositis by Mycoplasma hyorhinis
- PMID: 34069250
- PMCID: PMC8156107
- DOI: 10.3390/pathogens10050603
Altered Nasal Microbiota Composition Associated with Development of Polyserositis by Mycoplasma hyorhinis
Abstract
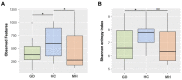
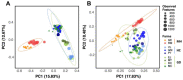
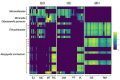
Fibrinous polyserositis in swine farming is a common pathological finding in nursery animals. The differential diagnosis of this finding should include Glaesserella parasuis (aetiological agent of Glässer's disease) and Mycoplasma hyorhinis, among others. These microorganisms are early colonizers of the upper respiratory tract of piglets. The composition of the nasal microbiota at weaning was shown to constitute a predisposing factor for the development of Glässer's disease. Here, we unravel the role of the nasal microbiota in the subsequent systemic infection by M. hyorhinis, and the similarities and differences with Glässer's disease. Nasal samples from farms with recurrent problems with polyserositis associated with M. hyorhinis (MH) or Glässer's disease (GD) were included in this study, together with healthy control farms (HC). Nasal swabs were taken from piglets in MH farms at weaning, before the onset of the clinical outbreaks, and were submitted to 16S rRNA gene amplicon sequencing (V3-V4 region). These sequences were analyzed together with sequences from similar samples previously obtained in GD and HC farms. Animals from farms with disease (MH and GD) had a nasal microbiota with lower diversity than those from the HC farms. However, the composition of the nasal microbiota of the piglets from these disease farms was different, suggesting that divergent microbiota imbalances may predispose the animals to the two systemic infections. We also found variants of the pathogens that were associated with the farms with the corresponding disease, highlighting the importance of studying the microbiome at strain-level resolution.
Keywords: 16S rRNA gene; Glässer’s disease; Mycoplasma hyorhinis; microbial diversity; nasal microbiota; porcine polyserositis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- McCormack U.M., Curião T., Buzoianu S.G., Prieto M.L., Ryan T., Varley P., Crispie F., Magowan E., Metzler-Zebeli B.U., Berry D., et al. Exploring a Possible Link between the Intestinal Microbiota and Feed Efficiency in Pigs. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00380-17. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

