Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale
- PMID: 34069261
- PMCID: PMC8156592
- DOI: 10.3390/ijms22105221
Genome-Wide Identification and Analysis of the APETALA2 (AP2) Transcription Factor in Dendrobium officinale
Abstract
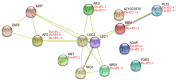
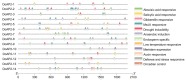
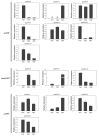
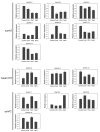
The APETALA2 (AP2) transcription factors (TFs) play crucial roles in regulating development in plants. However, a comprehensive analysis of the AP2 family members in a valuable Chinese herbal orchid, Dendrobium officinale, or in other orchids, is limited. In this study, the 14 DoAP2 TFs that were identified from the D. officinale genome and named DoAP2-1 to DoAP2-14 were divided into three clades: euAP2, euANT, and basalANT. The promoters of all DoAP2 genes contained cis-regulatory elements related to plant development and also responsive to plant hormones and stress. qRT-PCR analysis showed the abundant expression of DoAP2-2, DoAP2-5, DoAP2-7, DoAP2-8 and DoAP2-12 genes in protocorm-like bodies (PLBs), while DoAP2-3, DoAP2-4, DoAP2-6, DoAP2-9, DoAP2-10 and DoAP2-11 expression was strong in plantlets. In addition, the expression of some DoAP2 genes was down-regulated during flower development. These results suggest that DoAP2 genes may play roles in plant regeneration and flower development in D. officinale. Four DoAP2 genes (DoAP2-1 from euAP2, DoAP2-2 from euANT, and DoAP2-6 and DoAP2-11 from basal ANT) were selected for further analyses. The transcriptional activation of DoAP2-1, DoAP2-2, DoAP2-6 and DoAP2-11 proteins, which were localized in the nucleus of Arabidopsis thaliana mesophyll protoplasts, was further analyzed by a dual-luciferase reporter gene system in Nicotiana benthamiana leaves. Our data showed that pBD-DoAP2-1, pBD-DoAP2-2, pBD-DoAP2-6 and pBD-DoAP2-11 significantly repressed the expression of the LUC reporter compared with the negative control (pBD), suggesting that these DoAP2 proteins may act as transcriptional repressors in the nucleus of plant cells. Our findings on AP2 genes in D. officinale shed light on the function of AP2 genes in this orchid and other plant species.
Keywords: AP2 transcription factor; Dendrobium officinale; development; dual-luciferase reporter gene system; gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Sakuma Y., Liu Q., Dubouzet J.G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

