Ablation of Selenbp1 Alters Lipid Metabolism via the Pparα Pathway in Mouse Kidney
- PMID: 34069420
- PMCID: PMC8159118
- DOI: 10.3390/ijms22105334
Ablation of Selenbp1 Alters Lipid Metabolism via the Pparα Pathway in Mouse Kidney
Abstract
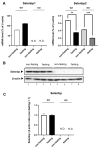
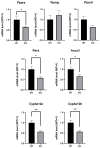
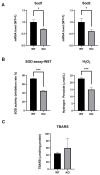
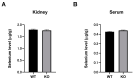
Selenium-binding protein 1 (Selenbp1) is a 2,3,7,8-tetrechlorodibenzo-p-dioxin inducible protein whose function is yet to be comprehensively elucidated. As the highly homologous isoform, Selenbp2, is expressed at low levels in the kidney, it is worthwhile comparing wild-type C57BL mice and Selenbp1-deficient mice under dioxin-free conditions. Accordingly, we conducted a mouse metabolomics analysis under non-dioxin-treated conditions. DNA microarray analysis was performed based on observed changes in lipid metabolism-related factors. The results showed fluctuations in the expression of numerous genes. Real-time RT-PCR confirmed the decreased expression levels of the cytochrome P450 4a (Cyp4a) subfamily, known to be involved in fatty acid ω- and ω-1 hydroxylation. Furthermore, peroxisome proliferator-activated receptor-α (Pparα) and retinoid-X-receptor-α (Rxrα), which form a heterodimer with Pparα to promote gene expression, were simultaneously reduced. This indicated that reduced Cyp4a expression was mediated via decreased Pparα and Rxrα. In line with this finding, increased levels of leukotrienes and prostaglandins were detected. Conversely, decreased hydrogen peroxide levels and reduced superoxide dismutase (SOD) activity supported the suppression of the renal expression of Sod1 and Sod2 in Selenbp1-deficient mice. Therefore, we infer that ablation of Selenbp1 elicits oxidative stress caused by increased levels of superoxide anions, which alters lipid metabolism via the Pparα pathway.
Keywords: Ppar; kidney; lipid metabolism; mouse; oxidative stress; peroxisome proliferator-activated receptor-alpha; selenium binding protein 1.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Elevated basal expression of liver peroxisomal beta-oxidation enzymes and CYP4A microsomal fatty acid omega-hydroxylase in STAT5b(-/-) mice: cross-talk in vivo between peroxisome proliferator-activated receptor and signal transducer and activator of transcription signaling pathways.Toxicol Appl Pharmacol. 2002 Jul 1;182(1):1-10. doi: 10.1006/taap.2002.9426. Toxicol Appl Pharmacol. 2002. PMID: 12127257
-
Peroxisome Proliferator Activated Receptor-α Association With Silent Information Regulator 1 Suppresses Cardiac Fatty Acid Metabolism in the Failing Heart.Circ Heart Fail. 2015 Nov;8(6):1123-32. doi: 10.1161/CIRCHEARTFAILURE.115.002216. Epub 2015 Oct 6. Circ Heart Fail. 2015. PMID: 26443578 Free PMC article.
-
Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression.Endocrinology. 2006 Mar;147(3):1508-16. doi: 10.1210/en.2005-1132. Epub 2005 Dec 15. Endocrinology. 2006. PMID: 16357043
-
Peroxisome proliferator-activated receptor alpha-mediated pathways are altered in hepatocyte-specific retinoid X receptor alpha-deficient mice.J Biol Chem. 2000 Sep 8;275(36):28285-90. doi: 10.1074/jbc.M000934200. J Biol Chem. 2000. PMID: 10866995
-
The role and regulation of the peroxisome proliferator activated receptor alpha in human liver.Biochimie. 2017 May;136:75-84. doi: 10.1016/j.biochi.2016.12.019. Epub 2017 Jan 8. Biochimie. 2017. PMID: 28077274 Review.
Cited by
-
RNA-Seq Analysis of Protection against Chronic Alcohol Liver Injury by Rosa roxburghii Fruit Juice (Cili) in Mice.Nutrients. 2022 May 9;14(9):1974. doi: 10.3390/nu14091974. Nutrients. 2022. PMID: 35565941 Free PMC article.
-
Hepatoprotective role of peroxisome proliferator-activated receptor-α in non-cancerous hepatic tissues following transcatheter arterial embolization.Open Life Sci. 2022 Aug 11;17(1):827-838. doi: 10.1515/biol-2022-0068. eCollection 2022. Open Life Sci. 2022. PMID: 36045714 Free PMC article.
-
Proteomic analysis of mouse liver lesions at all three stages of Echinococcus granulosus infection.PLoS Negl Trop Dis. 2024 Dec 3;18(12):e0012659. doi: 10.1371/journal.pntd.0012659. eCollection 2024 Dec. PLoS Negl Trop Dis. 2024. PMID: 39625960 Free PMC article.
-
Trace Element Selenium Effectively Alleviates Intestinal Diseases.Int J Mol Sci. 2021 Oct 28;22(21):11708. doi: 10.3390/ijms222111708. Int J Mol Sci. 2021. PMID: 34769138 Free PMC article. Review.
-
Ablation of Mouse Selenium-Binding Protein 1 and 2 Elevates LDL by Disruption of Cholesterol Efflux and Lipid Metabolism.Int J Mol Sci. 2025 Apr 3;26(7):3363. doi: 10.3390/ijms26073363. Int J Mol Sci. 2025. PMID: 40244197 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

