Contribution of Syndecans to the Cellular Entry of SARS-CoV-2
- PMID: 34069441
- PMCID: PMC8159090
- DOI: 10.3390/ijms22105336
Contribution of Syndecans to the Cellular Entry of SARS-CoV-2
Abstract
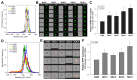
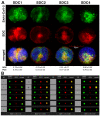
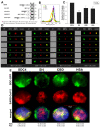
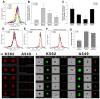
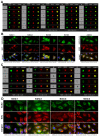
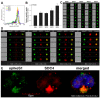
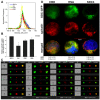
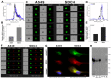
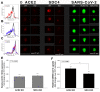
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel emerging pathogen causing an unprecedented pandemic in 21st century medicine. Due to the significant health and economic burden of the current SARS-CoV-2 outbreak, there is a huge unmet medical need for novel interventions effectively blocking SARS-CoV-2 infection. Unknown details of SARS-CoV-2 cellular biology hamper the development of potent and highly specific SARS-CoV-2 therapeutics. Angiotensin-converting enzyme-2 (ACE2) has been reported to be the primary receptor for SARS-CoV-2 cellular entry. However, emerging scientific evidence suggests the involvement of additional membrane proteins, such as heparan sulfate proteoglycans, in SARS-CoV-2 internalization. Here, we report that syndecans, the evolutionarily conserved family of transmembrane proteoglycans, facilitate the cellular entry of SARS-CoV-2. Among syndecans, the lung abundant syndecan-4 was the most efficient in mediating SARS-CoV-2 uptake. The S1 subunit of the SARS-CoV-2 spike protein plays a dominant role in the virus's interactions with syndecans. Besides the polyanionic heparan sulfate chains, other parts of the syndecan ectodomain, such as the cell-binding domain, also contribute to the interaction with SARS-CoV-2. During virus internalization, syndecans colocalize with ACE2, suggesting a jointly shared internalization pathway. Both ACE2 and syndecan inhibitors exhibited significant efficacy in reducing the cellular entry of SARS-CoV-2, thus supporting the complex nature of internalization. Data obtained on syndecan specific in vitro assays present syndecans as novel cellular targets of SARS-CoV-2 and offer molecularly precise yet simple strategies to overcome the complex nature of SARS-CoV-2 infection.
Keywords: SARS-CoV-2; cellular entry; coronaviruses; spike protein; syndecans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Syndecan-4 Is a Key Facilitator of the SARS-CoV-2 Delta Variant's Superior Transmission.Int J Mol Sci. 2022 Jan 12;23(2):796. doi: 10.3390/ijms23020796. Int J Mol Sci. 2022. PMID: 35054983 Free PMC article.
-
Biodistribution and Cellular Internalization of Inactivated SARS-CoV-2 in Wild-Type Mice.Int J Mol Sci. 2022 Jul 9;23(14):7609. doi: 10.3390/ijms23147609. Int J Mol Sci. 2022. PMID: 35886958 Free PMC article.
-
The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target.Tumour Biol. 2021;43(1):177-196. doi: 10.3233/TUB-200084. Tumour Biol. 2021. PMID: 34420993 Review.
-
Potential therapeutic approaches for the early entry of SARS-CoV-2 by interrupting the interaction between the spike protein on SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2).Biochem Pharmacol. 2021 Oct;192:114724. doi: 10.1016/j.bcp.2021.114724. Epub 2021 Aug 8. Biochem Pharmacol. 2021. PMID: 34371003 Free PMC article. Review.
-
Mutual Inhibition of Antithrombin III and SARS-CoV-2 Cellular Attachment to Syndecans: Implications for COVID-19 Treatment and Vaccination.Int J Mol Sci. 2024 Jul 9;25(14):7534. doi: 10.3390/ijms25147534. Int J Mol Sci. 2024. PMID: 39062776 Free PMC article.
Cited by
-
Antiviral strategies should focus on stimulating the biosynthesis of heparan sulfates, not their inhibition.Life Sci. 2021 Jul 15;277:119508. doi: 10.1016/j.lfs.2021.119508. Epub 2021 Apr 15. Life Sci. 2021. PMID: 33865880 Free PMC article. Review.
-
Apolipoprotein E and viral infection: Risks and Mechanisms.Mol Ther Nucleic Acids. 2023 Jul 28;33:529-542. doi: 10.1016/j.omtn.2023.07.031. eCollection 2023 Sep 12. Mol Ther Nucleic Acids. 2023. PMID: 37588688 Free PMC article. Review.
-
Impact of a Functional Dairy Powder and Its Primary Component on the Growth of Pathogenic and Probiotic Gut Bacteria and Human Coronavirus 229E.Int J Mol Sci. 2024 Aug 29;25(17):9353. doi: 10.3390/ijms25179353. Int J Mol Sci. 2024. PMID: 39273301 Free PMC article.
-
Endocytic Pathways Unveil the Role of Syndecans in the Seeding and Spreading of Pathological Protein Aggregates: Insights into Neurodegenerative Disorders.Int J Mol Sci. 2025 Apr 24;26(9):4037. doi: 10.3390/ijms26094037. Int J Mol Sci. 2025. PMID: 40362276 Free PMC article. Review.
-
SARS-CoV-2 Lurking and Lingering in the Depths of Lungs.Am J Respir Crit Care Med. 2024 Apr 1;209(7):779-780. doi: 10.1164/rccm.202401-0067ED. Am J Respir Crit Care Med. 2024. PMID: 38324719 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

