Iron-Bound Lipocalin-2 Protects Renal Cell Carcinoma from Ferroptosis
- PMID: 34069743
- PMCID: PMC8161288
- DOI: 10.3390/metabo11050329
Iron-Bound Lipocalin-2 Protects Renal Cell Carcinoma from Ferroptosis
Abstract
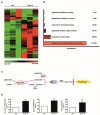
While the importance of the iron-load of lipocalin-2 (Lcn-2) in promoting tumor progression is widely appreciated, underlying molecular mechanisms largely remain elusive. Considering its role as an iron-transporter, we aimed at clarifying iron-loaded, holo-Lcn-2 (hLcn-2)-dependent signaling pathways in affecting renal cancer cell viability. Applying RNA sequencing analysis in renal CAKI1 tumor cells to explore highly upregulated molecular signatures in response to hLcn-2, we identified a cluster of genes (SLC7A11, GCLM, GLS), which are implicated in regulating ferroptosis. Indeed, hLcn-2-stimulated cells are protected from erastin-induced ferroptosis. We also noticed a rapid increase in reactive oxygen species (ROS) with subsequent activation of the antioxidant Nrf2 pathway. However, knocking down Nrf2 by siRNA was not sufficient to induce erastin-dependent ferroptotic cell death in hLcn-2-stimulated tumor cells. In contrast, preventing oxidative stress through N-acetyl-l-cysteine (NAC) supplementation was still able to induce erastin-dependent ferroptotic cell death in hLcn-2-stimulated tumor cells. Besides an oxidative stress response, we noticed activation of the integrated stress response (ISR), shown by enhanced phosphorylation of eIF-2α and induction of ATF4 after hLcn-2 addition. ATF4 knockdown as well as inhibition of the ISR sensitized hLcn-2-treated renal tumor cells to ferroptosis, thus linking the ISR to pro-tumor characteristics of hLcn-2. Our study provides mechanistic details to better understand tumor pro-survival pathways initiated by iron-loaded Lcn-2.
Keywords: ISR; Nrf2; ROS; SLC7A11; erastin; ferroptosis; iron; lipocalin-2; p-eIF2α.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells.Cell Death Dis. 2020 Sep 14;11(9):751. doi: 10.1038/s41419-020-02939-3. Cell Death Dis. 2020. PMID: 32929075 Free PMC article.
-
MicroRNA-214-3p enhances erastin-induced ferroptosis by targeting ATF4 in hepatoma cells.J Cell Physiol. 2020 Jul;235(7-8):5637-5648. doi: 10.1002/jcp.29496. Epub 2020 Jan 21. J Cell Physiol. 2020. PMID: 31960438
-
Knockdown of ferroportin accelerates erastin-induced ferroptosis in neuroblastoma cells.Eur Rev Med Pharmacol Sci. 2018 Jun;22(12):3826-3836. doi: 10.26355/eurrev_201806_15267. Eur Rev Med Pharmacol Sci. 2018. PMID: 29949159
-
Ferroptosis: process and function.Cell Death Differ. 2016 Mar;23(3):369-79. doi: 10.1038/cdd.2015.158. Epub 2016 Jan 22. Cell Death Differ. 2016. PMID: 26794443 Free PMC article. Review.
-
The Role of Erastin in Ferroptosis and Its Prospects in Cancer Therapy.Onco Targets Ther. 2020 Jun 11;13:5429-5441. doi: 10.2147/OTT.S254995. eCollection 2020. Onco Targets Ther. 2020. PMID: 32606760 Free PMC article. Review.
Cited by
-
Bioinformatic analysis of the LCN2-SLC22A17-MMP9 network in cancer: The role of DNA methylation in the modulation of tumor microenvironment.Front Cell Dev Biol. 2022 Sep 21;10:945586. doi: 10.3389/fcell.2022.945586. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36211450 Free PMC article.
-
Binding to Iron Quercetin Complexes Increases the Antioxidant Capacity of the Major Birch Pollen Allergen Bet v 1 and Reduces Its Allergenicity.Antioxidants (Basel). 2022 Dec 26;12(1):42. doi: 10.3390/antiox12010042. Antioxidants (Basel). 2022. PMID: 36670905 Free PMC article.
-
Lipocalin-2 and intestinal diseases.World J Gastroenterol. 2024 Dec 14;30(46):4864-4879. doi: 10.3748/wjg.v30.i46.4864. World J Gastroenterol. 2024. PMID: 39679305 Free PMC article. Review.
-
Targeting ferroptosis in renal cell carcinoma: Potential mechanisms and novel therapeutics.Heliyon. 2023 Jul 21;9(8):e18504. doi: 10.1016/j.heliyon.2023.e18504. eCollection 2023 Aug. Heliyon. 2023. PMID: 37554789 Free PMC article. Review.
-
LncRNA TMPO-AS1 facilitates cervical cancer cell tumorigenesis and ferroptosis resistance via interaction with LCN2.Sci Rep. 2025 Jul 2;15(1):22604. doi: 10.1038/s41598-025-07213-0. Sci Rep. 2025. PMID: 40594740 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

