A Self-Priming Microfluidic Chip with Cushion Chambers for Easy Digital PCR
- PMID: 34069758
- PMCID: PMC8155915
- DOI: 10.3390/bios11050158
A Self-Priming Microfluidic Chip with Cushion Chambers for Easy Digital PCR
Abstract
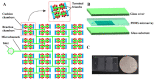
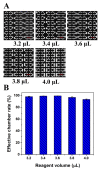
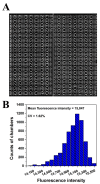
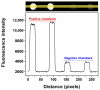
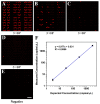
A polydimethylsiloxane (PDMS)-based self-priming microfluidic chip with cushion chambers is presented in this study for robust and easy-operation digital polymerase chain reaction (dPCR). The chip has only one inlet and can partition samples autonomously through negative pressure, provided by a de-gassed PDMS layer with a multi-level vertical branching microchannel design. Meanwhile, cushion chambers make the chip capable of very robust use for sample partitioning. Finally, the proposed microfluidic chip showed excellent performance in the absolute quantification of a target gene by performing quantitative detection of a 10-fold serial dilution DNA template. Owing to its characteristics of easy operation, low cost, and high robustness, the proposed dPCR chip is expected to further promote the extensive application of digital PCR, especially in resource-limited settings.
Keywords: cushion chambers; digital PCR; microfluidic chip; self-priming.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Vendrell J.A., Mazieres J., Senal R., Rouquette I., Quantin X., Pujol J.L., Roch B., Bouidioua A., Godreuil S., Coyaud E., et al. Ultra-sensitive EGFR (T790M) detection as an independent prognostic marker for lung cancer patients harboring EGFR (del19) mutations and treated with first-generation TKIs. Clin. Cancer Res. 2019;25:4280–4289. doi: 10.1158/1078-0432.CCR-18-2683. - DOI - PubMed
-
- Laurent-Puig P., Pekin D., Normand C., Kotsopoulos S.K., Nizard P., Perez-Toralla K., Rowell R., Olson J., Srinivasan P., Le Corre D., et al. Clinical relevance of KRAS-mutated subclones detected with picodroplet digital PCR in advanced colorectal cancer treated with anti-EGFR therapy. Clin. Cancer Res. 2015;21:1087–1097. doi: 10.1158/1078-0432.CCR-14-0983. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

