Genome-Wide Association Study Reveals Genomic Regions Associated with Fusarium Wilt Resistance in Common Bean
- PMID: 34069884
- PMCID: PMC8157364
- DOI: 10.3390/genes12050765
Genome-Wide Association Study Reveals Genomic Regions Associated with Fusarium Wilt Resistance in Common Bean
Abstract
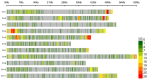
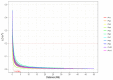
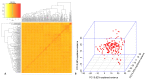
Fusarium wilt (Fusarium oxysporum f. sp. phaseoli, Fop) is one of the main fungal soil diseases in common bean. The aim of the present study was to identify genomic regions associated with Fop resistance through genome-wide association studies (GWAS) in a Mesoamerican Diversity Panel (MDP) and to identify potential common bean sources of Fop's resistance. The MDP was genotyped with BARCBean6K_3BeadChip and evaluated for Fop resistance with two different monosporic strains using the root-dip method. Disease severity rating (DSR) and the area under the disease progress curve (AUDPC), at 21 days after inoculation (DAI), were used for GWAS performed with FarmCPU model. The p-value of each SNP was determined by resampling method and Bonferroni test. For UFV01 strain, two significant single nucleotide polymorphisms (SNPs) were mapped on the Pv05 and Pv11 for AUDPC, and the same SNP (ss715648096) on Pv11 was associated with AUDPC and DSR. Another SNP, mapped on Pv03, showed significance for DSR. Regarding IAC18001 strain, significant SNPs on Pv03, Pv04, Pv05, Pv07 and on Pv01, Pv05, and Pv10 were observed. Putative candidate genes related to nucleotide-binding sites and carboxy-terminal leucine-rich repeats were identified. The markers may be important future tools for genomic selection to Fop disease resistance in beans.
Keywords: Fusarium oxysporum f. sp. phaseoli; Phaseolus vulgaris L.; SNP markers; disease resistance; molecular breeding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
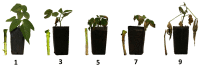
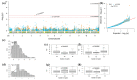
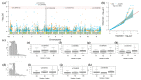
References
-
- Bitocchi E., Bellucci E., Giardini A., Rau D., Rodriguez M., Biagetti E., Santilocchi R., Spagnoletti Zeuli P., Gioia T., Logozzo G., et al. Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 2013;197:300–313. doi: 10.1111/j.1469-8137.2012.04377.x. - DOI - PubMed
-
- De Ron A.M., Santalla M. Brenner’s Encyclopedia of Genetics. 2nd ed. Elsevier Inc.; Amsterdam, The Netherlands: 2013. Phaseolus vulgaris (Beans) pp. 290–292.
-
- Singh S.P., Gepts P., Debouck D.G. Races of common bean (Phaseolus vulgaris, Fabaceae) Econ. Bot. 1991;45:379–396. doi: 10.1007/BF02887079. - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

