Prediction and Evolution of the Molecular Fitness of SARS-CoV-2 Variants: Introducing SpikePro
- PMID: 34070055
- PMCID: PMC8158131
- DOI: 10.3390/v13050935
Prediction and Evolution of the Molecular Fitness of SARS-CoV-2 Variants: Introducing SpikePro
Abstract
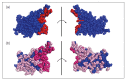
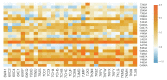
The understanding of the molecular mechanisms driving the fitness of the SARS-CoV-2 virus and its mutational evolution is still a critical issue. We built a simplified computational model, called SpikePro, to predict the SARS-CoV-2 fitness from the amino acid sequence and structure of the spike protein. It contains three contributions: the inter-human transmissibility of the virus predicted from the stability of the spike protein, the infectivity computed in terms of the affinity of the spike protein for the ACE2 receptor, and the ability of the virus to escape from the human immune response based on the binding affinity of the spike protein for a set of neutralizing antibodies. Our model reproduces well the available experimental, epidemiological and clinical data on the impact of variants on the biophysical characteristics of the virus. For example, it is able to identify circulating viral strains that, by increasing their fitness, recently became dominant at the population level. SpikePro is a useful, freely available instrument which predicts rapidly and with good accuracy the dangerousness of new viral strains. It can be integrated and play a fundamental role in the genomic surveillance programs of the SARS-CoV-2 virus that, despite all the efforts, remain time-consuming and expensive.
Keywords: COVID-19; SARS-CoV-2; deep mutagenesis; immune escape; protein binding affinity; protein stability; spike protein variants; viral fitness.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
SpikePro: a webserver to predict the fitness of SARS-CoV-2 variants.Bioinformatics. 2022 Sep 15;38(18):4418-4419. doi: 10.1093/bioinformatics/btac517. Bioinformatics. 2022. PMID: 35861514
-
Risk of rapid evolutionary escape from biomedical interventions targeting SARS-CoV-2 spike protein.PLoS One. 2021 Apr 28;16(4):e0250780. doi: 10.1371/journal.pone.0250780. eCollection 2021. PLoS One. 2021. PMID: 33909660 Free PMC article.
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Angiotensin-Converting Enzyme 2 (ACE2) in the Pathogenesis of ARDS in COVID-19.Front Immunol. 2021 Dec 22;12:732690. doi: 10.3389/fimmu.2021.732690. eCollection 2021. Front Immunol. 2021. PMID: 35003058 Free PMC article. Review.
-
SARS-CoV-2 Entry Related Viral and Host Genetic Variations: Implications on COVID-19 Severity, Immune Escape, and Infectivity.Int J Mol Sci. 2021 Mar 17;22(6):3060. doi: 10.3390/ijms22063060. Int J Mol Sci. 2021. PMID: 33802729 Free PMC article. Review.
Cited by
-
Biophysical principles predict fitness of SARS-CoV-2 variants.bioRxiv [Preprint]. 2024 Jan 22:2023.07.23.549087. doi: 10.1101/2023.07.23.549087. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2024 Jun 4;121(23):e2314518121. doi: 10.1073/pnas.2314518121. PMID: 37577536 Free PMC article. Updated. Preprint.
-
An integrated understanding of the evolutionary and structural features of the SARS-CoV-2 spike receptor binding domain (RBD).Int J Biol Macromol. 2022 Sep 30;217:492-505. doi: 10.1016/j.ijbiomac.2022.07.022. Epub 2022 Jul 13. Int J Biol Macromol. 2022. PMID: 35841961 Free PMC article.
-
In silico genomic surveillance by CoVerage predicts and characterizes SARS-CoV-2 variants of interest.Nat Commun. 2025 Jul 8;16(1):6281. doi: 10.1038/s41467-025-60231-4. Nat Commun. 2025. PMID: 40628697 Free PMC article.
-
Early warning of emerging infectious diseases based on multimodal data.Biosaf Health. 2023 Jun 7;5(4):193-203. doi: 10.1016/j.bsheal.2023.05.006. Online ahead of print. Biosaf Health. 2023. PMID: 37362865 Free PMC article. Review.
-
AlphaFold2-Enabled Atomistic Modeling of Structure, Conformational Ensembles, and Binding Energetics of the SARS-CoV-2 Omicron BA.2.86 Spike Protein with ACE2 Host Receptor and Antibodies: Compensatory Functional Effects of Binding Hotspots in Modulating Mechanisms of Receptor Binding and Immune Escape.J Chem Inf Model. 2024 Mar 11;64(5):1657-1681. doi: 10.1021/acs.jcim.3c01857. Epub 2024 Feb 19. J Chem Inf Model. 2024. PMID: 38373700 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

