A Systematic Evaluation of Semispecific Peptide Search Parameter Enables Identification of Previously Undescribed N-Terminal Peptides and Conserved Proteolytic Processing in Cancer Cell Lines
- PMID: 34070654
- PMCID: PMC8162549
- DOI: 10.3390/proteomes9020026
A Systematic Evaluation of Semispecific Peptide Search Parameter Enables Identification of Previously Undescribed N-Terminal Peptides and Conserved Proteolytic Processing in Cancer Cell Lines
Abstract
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) has become the most commonly used technique in explorative proteomic research. A variety of open-source tools for peptide-spectrum matching have become available. Most analyses of explorative MS data are performed using conventional settings, such as fully specific enzymatic constraints. Here we evaluated the impact of the fragment mass tolerance in combination with the enzymatic constraints on the performance of three search engines. Three open-source search engines (Myrimatch, X! Tandem, and MSGF+) were evaluated concerning the suitability in semi- and unspecific searches as well as the importance of accurate fragment mass spectra in non-specific peptide searches. We then performed a semispecific reanalysis of the published NCI-60 deep proteome data applying the most suited parameters. Semi- and unspecific LC-MS/MS data analyses particularly benefit from accurate fragment mass spectra while this effect is less pronounced for conventional, fully specific peptide-spectrum matching. Search speed differed notably between the three search engines for semi- and non-specific peptide-spectrum matching. Semispecific reanalysis of NCI-60 proteome data revealed hundreds of previously undescribed N-terminal peptides, including cases of proteolytic processing or likely alternative translation start sites, some of which were ubiquitously present in all cell lines of the reanalyzed panel. Highly accurate MS2 fragment data in combination with modern open-source search algorithms enable the confident identification of semispecific peptides from large proteomic datasets. The identification of previously undescribed N-terminal peptides in published studies highlights the potential of future reanalysis and data mining in proteomic datasets.
Keywords: NCI-60 reanalysis; endogenous proteolysis; fragment mass tolerance; mass spectrometry; semispecific peptide search.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures


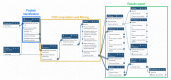
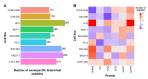
References
-
- Föll M.C., Fahrner M., Gretzmeier C., Thoma K., Biniossek M.L., Kiritsi D., Meiss F., Schilling O., Nyström A., Kern J.S. Identification of tissue damage, extracellular matrix remodeling and bacterial challenge as common mechanisms associated with high-risk cutaneous squamous cell carcinomas. Matrix Biol. 2018;66:1–21. doi: 10.1016/j.matbio.2017.11.004. - DOI - PubMed
-
- Oria V., Bronsert P., Thomsen A., Föll M., Zamboglou C., Hannibal L., Behringer S., Biniossek M., Schreiber C., Grosu A., et al. Proteome Profiling of Primary Pancreatic Ductal Adenocarcinomas Undergoing Additive Chemoradiation Link ALDH1A1 to Early Local Recurrence and Chemoradiation Resistance. Transl. Oncol. 2018;11:1307–1322. doi: 10.1016/j.tranon.2018.08.001. - DOI - PMC - PubMed
-
- Müller A.-K., Föll M., Heckelmann B., Kiefer S., Werner M., Schilling O., Biniossek M.L., Jilg C.A., Drendel V. Proteomic Characterization of Prostate Cancer to Distinguish Nonmetastasizing and Metastasizing Primary Tumors and Lymph Node Metastases. Neoplasia. 2018;20:140–151. doi: 10.1016/j.neo.2017.10.009. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

