Development of Real-Time and Conventional PCR Assays for Identifying a Newly Named Species of Root-Lesion Nematode (Pratylenchus dakotaensis) on Soybean
- PMID: 34070906
- PMCID: PMC8197872
- DOI: 10.3390/ijms22115872
Development of Real-Time and Conventional PCR Assays for Identifying a Newly Named Species of Root-Lesion Nematode (Pratylenchus dakotaensis) on Soybean
Abstract
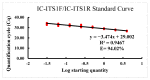
A rapid and accurate PCR-based method was developed in this study for detecting and identifying a new species of root-lesion nematode (Pratylenchus dakotaensis) recently discovered in a soybean field in North Dakota, USA. Species-specific primers, targeting the internal transcribed spacer region of ribosomal DNA, were designed to be used in both conventional and quantitative real-time PCR assays for identification of P.dakotaensis. The specificity of the primers was evaluated in silico analysis and laboratory PCR experiments. Results showed that only P.dakotaensis DNA was exclusively amplified in conventional and real-time PCR assays but none of the DNA from other control species were amplified. Detection sensitivity analysis revealed that the conventional PCR was able to detect an equivalent to 1/8 of the DNA of a single nematode whereas real-time PCR detected an equivalent to 1/32 of the DNA of a single nematode. According to the generated standard curve the amplification efficiency of the primers in real-time PCR was 94% with a R2 value of 0.95 between quantification cycle number and log number of P.dakotaensis. To validate the assays to distinguish P.dakotaensis from other Pratylenchus spp. commonly detected in North Dakota soybean fields, 20 soil samples collected from seven counties were tested. The PCR assays amplified the DNA of P.dakotaensis and discriminated it from other Pratylenchus spp. present in North Dakota soybean fields. This is the first report of a species-specific and rapid PCR detection method suitable for use in diagnostic and research laboratories for the detection of P.dakotaensis.
Keywords: DNA; ITS rDNA; Pratylenchus dakotaensis; conventional PCR; detection; endomigratory; identification; in silico analysis; real-time PCR; root-lesion nematode.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Development of a Recombinase Polymerase Amplification Assay for Rapid Detection of the New Root-Lesion Nematode Pratylenchus dakotaensis on Soybean.Plant Dis. 2025 Mar;109(3):603-614. doi: 10.1094/PDIS-05-24-1133-RE. Epub 2025 Feb 28. Plant Dis. 2025. PMID: 39320378
-
Detection and quantification of Pratylenchus thornei in DNA extracted from soil using real-time PCR.Phytopathology. 2012 Jan;102(1):14-22. doi: 10.1094/PHYTO-03-11-0093. Phytopathology. 2012. PMID: 21879792
-
Population Development of the Root-Lesion Nematode Pratylenchus dakotaensis on Soybean Cultivars.Plant Dis. 2022 Aug;106(8):2117-2126. doi: 10.1094/PDIS-11-21-2444-RE. Epub 2022 Jul 19. Plant Dis. 2022. PMID: 35147453
-
Morphological and Molecular Characterization of Pratylenchus dakotaensis n. sp. (Nematoda: Pratylenchidae), a New Root-Lesion Nematode Species on Soybean in North Dakota, USA.Plants (Basel). 2021 Jan 17;10(1):168. doi: 10.3390/plants10010168. Plants (Basel). 2021. PMID: 33477266 Free PMC article.
-
Advances in Understanding the Molecular Mechanisms of Root Lesion Nematode Host Interactions.Annu Rev Phytopathol. 2016 Aug 4;54:253-78. doi: 10.1146/annurev-phyto-080615-100257. Epub 2016 Jan 1. Annu Rev Phytopathol. 2016. PMID: 27296144 Review.
Cited by
-
Rapid and Accurate Detection of Gnomoniopsis smithogilvyi the Causal Agent of Chestnut Rot, through an Internally Controlled Multiplex PCR Assay.Pathogens. 2022 Aug 12;11(8):907. doi: 10.3390/pathogens11080907. Pathogens. 2022. PMID: 36015028 Free PMC article.
-
Prevalence of pest nematodes associated with soybean (Glycine max) in Wisconsin from 1998 to 2021.J Nematol. 2024 Mar 30;55(1):20230053. doi: 10.2478/jofnem-2023-0053. eCollection 2023 Feb. J Nematol. 2024. PMID: 38558769 Free PMC article.
-
Molecular Profiling and Precise Diagnosis of Pratylenchus penetrans Infestation in Soil: A qPCR-Based Molecular Approach.Plant Pathol J. 2025 Jun;41(3):330-340. doi: 10.5423/PPJ.OA.11.2024.0181. Epub 2025 Jun 1. Plant Pathol J. 2025. PMID: 40468880 Free PMC article.
References
-
- Sasser J.N., Freckman D.W. A world perspective on nematology: The role of the society. In: Veech J.A., Dickerson D.W., editors. Vistas on Nematology: A Commemoration of the Twenty-Fifth Anniversary of the Society of Nematologists. 1st ed. Society of Nematologists; Lakeland, FL, USA: 1987. pp. 7–14.
-
- Castillo P., Vovlas N. Pratylenchus (Nematoda: Pratylenchidae): Diagnosis, biology, pathogenicity and management. Nematol. Monogr. Perspect. 2007;6:1–543. doi: 10.1163/ej.9789004155640.i-523. - DOI
-
- Davis E.L., MacGuidwin A.E. Lesion nematode disease. Plant Health Instr. 2000 doi: 10.1094/PHI-I-2000-1030-02. - DOI
-
- Taylor S.P., Vanstone V.A., Ware A.H., McKay A.C., Szot D., Russ M.H. Measuring yield loss in cereals caused by root-lesion nematodes (Pratylenchus Neglectus and P. Thornei) with and without nematicide. Aust. J. Agric. Res. 1999;50:617–622. doi: 10.1071/A98103. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

