Bacterial Diversity and Community Structure of a Municipal Solid Waste Landfill: A Source of Lignocellulolytic Potential
- PMID: 34071172
- PMCID: PMC8228822
- DOI: 10.3390/life11060493
Bacterial Diversity and Community Structure of a Municipal Solid Waste Landfill: A Source of Lignocellulolytic Potential
Abstract
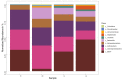
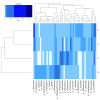
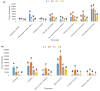
Omics have given rise to research on sparsely studied microbial communities such as the landfill, lignocellulolytic microorganisms and enzymes. The bacterial diversity of Municipal Solid Waste sediments was determined using the illumina MiSeq system after DNA extraction and Polymerase chain reactions. Data analysis was used to determine the community's richness, diversity, and correlation with environmental factors. Physicochemical studies revealed sites with mesophilic and thermophilic temperature ranges and a mixture of acidic and alkaline pH values. Temperature and moisture content showed the highest correlation with the bacteria community. The bacterial analysis of the community DNA revealed 357,030 effective sequences and 1891 operational taxonomic units (OTUs) assigned. Forty phyla were found, with the dominant phyla Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidota, while Aerococcus, Stenotrophomonas, and Sporosarcina were the dominant species. PICRUSt provided insight on community's metabolic function, which was narrowed down to search for lignocellulolytic enzymes' function. Cellulase, xylanase, esterase, and peroxidase were gene functions inferred from the data. This article reports on the first phylogenetic analysis of the Pulau Burung landfill bacterial community. These results will help to improve the understanding of organisms dominant in the landfill and the corresponding enzymes that contribute to lignocellulose breakdown.
Keywords: bacteria; biodiversity; landfill; lignocellulolytic bacteria; lignocellulolytic enzyme; lignocellulose biomass; metagenomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Metagenomics profiling for assessing microbial diversity in both active and closed landfills.Sci Total Environ. 2018 Mar;616-617:269-278. doi: 10.1016/j.scitotenv.2017.10.266. Epub 2017 Nov 5. Sci Total Environ. 2018. PMID: 29117585
-
Structure and diversity of bacterial communities in two large sanitary landfills in China as revealed by high-throughput sequencing (MiSeq).Waste Manag. 2017 May;63:41-48. doi: 10.1016/j.wasman.2016.07.047. Epub 2016 Aug 8. Waste Manag. 2017. PMID: 27515184
-
Lignocellulose-Degrading Microbial Communities in Landfill Sites Represent a Repository of Unexplored Biomass-Degrading Diversity.mSphere. 2017 Aug 2;2(4):e00300-17. doi: 10.1128/mSphere.00300-17. eCollection 2017 Jul-Aug. mSphere. 2017. PMID: 28776044 Free PMC article.
-
Community diversity metrics, interactions, and metabolic functions of bacteria associated with municipal solid waste landfills at different maturation stages.Microbiologyopen. 2021 Jan;10(1):e1118. doi: 10.1002/mbo3.1118. Epub 2020 Dec 12. Microbiologyopen. 2021. PMID: 33314739 Free PMC article.
-
Microbial community composition and metabolic functions in landfill leachate from different landfills of China.Sci Total Environ. 2021 May 1;767:144861. doi: 10.1016/j.scitotenv.2020.144861. Epub 2020 Dec 31. Sci Total Environ. 2021. PMID: 33422962
Cited by
-
Uncontrolled Post-Industrial Landfill-Source of Metals, Potential Toxic Compounds, Dust, and Pathogens in Environment-A Case Study.Molecules. 2024 Mar 27;29(7):1496. doi: 10.3390/molecules29071496. Molecules. 2024. PMID: 38611776 Free PMC article.
-
The Potential of Aloe vera and Opuntia ficus-indica Extracts as Biobased Agents for the Conservation of Cultural Heritage Metals.Metabolites. 2025 Jun 10;15(6):386. doi: 10.3390/metabo15060386. Metabolites. 2025. PMID: 40559410 Free PMC article.
-
Assessment of Physicochemical, Microbiological and Toxicological Hazards at an Illegal Landfill in Central Poland.Int J Environ Res Public Health. 2022 Apr 15;19(8):4826. doi: 10.3390/ijerph19084826. Int J Environ Res Public Health. 2022. PMID: 35457694 Free PMC article.
-
A Microcosm Model for the Study of Microbial Community Shift and Carbon Emission from Landfills.Indian J Microbiol. 2022 Jun;62(2):195-203. doi: 10.1007/s12088-021-00995-7. Epub 2022 Jan 11. Indian J Microbiol. 2022. PMID: 35462719 Free PMC article.
-
Optimization and comparative study of Bacillus consortia for cellulolytic potential and cellulase enzyme activity.Open Life Sci. 2025 Mar 6;20(1):20251066. doi: 10.1515/biol-2025-1066. eCollection 2025. Open Life Sci. 2025. PMID: 40059875 Free PMC article.
References
-
- Abdel-Shafy H.I., Mansour M.S.M. Solid waste issue: Sources, composition, disposal, recycling, and valorization. Egypt. J. Pet. 2018;27:1275–1290. doi: 10.1016/j.ejpe.2018.07.003. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

