Quantitative Detection of Bifidobacterium longum Strains in Feces Using Strain-Specific Primers
- PMID: 34071208
- PMCID: PMC8227663
- DOI: 10.3390/microorganisms9061159
Quantitative Detection of Bifidobacterium longum Strains in Feces Using Strain-Specific Primers
Abstract
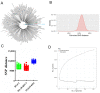
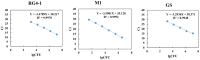
We adopted a bioinformatics-based technique to identify strain-specific markers, which were then used to quantify the abundances of three distinct B. longum sup. longum strains in fecal samples of humans and mice. A pangenome analysis of 205 B. longum sup. longum genomes revealed the accumulation of considerable strain-specific genes within this species; specifically, 28.7% of the total identified genes were strain-specific. We identified 32, 14, and 49 genes specific to B. longum sup. longum RG4-1, B. longum sup. longum M1-20-R01-3, and B. longum sup. longum FGSZY6M4, respectively. After performing an in silico validation of these strain-specific markers using a nucleotide BLAST against both the B. longum sup. longum genome database and an NR/NT database, RG4-1_01874 (1331 bp), M1-20-R01-3_00324 (1745 bp), and FGSZY6M4_01477 (1691 bp) were chosen as target genes for strain-specific quantification. The specificities of the qPCR primers were validated against 47 non-target microorganisms and fecal baseline microbiota to ensure that they produced no PCR amplification products. The performance of the qPCR primer-based analysis was further assessed using fecal samples. After oral administration, the target B. longum strains appeared to efficiently colonize both the human and mouse guts, with average population levels of >108 CFU/g feces. The bioinformatics pipeline proposed here can be applied to the quantification of various bacterial species.
Keywords: B. longum sup. longum; Roary; bioinformatics; gut colonization; probiotics; strain-specific qualification.
Conflict of interest statement
There are no conflict of interest to declare.
Figures


Similar articles
-
tuf Gene Sequence Variation in Bifidobacterium longum subsp. infantis Detected in the Fecal Microbiota of Chinese Infants.Appl Environ Microbiol. 2018 Jun 18;84(13):e00336-18. doi: 10.1128/AEM.00336-18. Print 2018 Jul 1. Appl Environ Microbiol. 2018. PMID: 29703739 Free PMC article.
-
Long-term colonization exceeding six years from early infancy of Bifidobacterium longum subsp. longum in human gut.BMC Microbiol. 2018 Dec 12;18(1):209. doi: 10.1186/s12866-018-1358-6. BMC Microbiol. 2018. PMID: 30541439 Free PMC article.
-
Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets.PeerJ. 2017 May 25;5:e3375. doi: 10.7717/peerj.3375. eCollection 2017. PeerJ. 2017. PMID: 28560114 Free PMC article.
-
Safety and intestinal microbiota modulation by the exopolysaccharide-producing strains Bifidobacterium animalis IPLA R1 and Bifidobacterium longum IPLA E44 orally administered to Wistar rats.Int J Food Microbiol. 2011 Jan 5;144(3):342-51. doi: 10.1016/j.ijfoodmicro.2010.10.016. Int J Food Microbiol. 2011. PMID: 21078530
-
Exploring the Dose-Effect Relationship of Bifidobacterium longum in Relieving Loperamide Hydrochloride-Induced Constipation in Rats through Colon-Released Capsules.Int J Mol Sci. 2023 Apr 1;24(7):6585. doi: 10.3390/ijms24076585. Int J Mol Sci. 2023. PMID: 37047557 Free PMC article.
Cited by
-
Bifidobacterium breve and Bifidobacterium longum Attenuate Choline-Induced Plasma Trimethylamine N-Oxide Production by Modulating Gut Microbiota in Mice.Nutrients. 2022 Mar 14;14(6):1222. doi: 10.3390/nu14061222. Nutrients. 2022. PMID: 35334879 Free PMC article.
-
Colonized Niche, Evolution and Function Signatures of Bifidobacterium pseudolongum within Bifidobacterial Genus.Foods. 2021 Sep 27;10(10):2284. doi: 10.3390/foods10102284. Foods. 2021. PMID: 34681333 Free PMC article.
-
Maternal-infant probiotic transmission mitigates early-life stress-induced autism in mice.Gut Microbes. 2025 Dec;17(1):2456584. doi: 10.1080/19490976.2025.2456584. Epub 2025 Feb 11. Gut Microbes. 2025. PMID: 39931863 Free PMC article.
-
Bacteroides vulgatus Ameliorates Lipid Metabolic Disorders and Modulates Gut Microbial Composition in Hyperlipidemic Rats.Microbiol Spectr. 2023 Feb 14;11(1):e0251722. doi: 10.1128/spectrum.02517-22. Epub 2023 Jan 10. Microbiol Spectr. 2023. PMID: 36625637 Free PMC article.
-
Bacteroides vulgatus attenuates experimental mice colitis through modulating gut microbiota and immune responses.Front Immunol. 2022 Nov 30;13:1036196. doi: 10.3389/fimmu.2022.1036196. eCollection 2022. Front Immunol. 2022. PMID: 36531989 Free PMC article.
References
-
- Krumbeck J.A., Maldonado-Gomez M.X., Martínez I., Frese S.A., Burkey T.E., Rasineni K., Ramer-Tait A.E., Harris E.N., Hutkins R.W., Walter J. In Vivo Selection To Identify Bacterial Strains with Enhanced Ecological Performance in Synbiotic Applications. Appl. Environ. Microbiol. 2015;81:2455–2465. doi: 10.1128/AEM.03903-14. - DOI - PMC - PubMed
Grants and funding
- No. 31820103010 and No. 31871773/the National Natural Science Foundation of China Program
- 2018DB002/Projects of Innovation and Development Pillar Program for Key Industries in Southern Xinjiang of Xinjiang Production and Construction Corps
- No. 2018YFC1604206/National Key Research and Development Project
- JUFSTR20180102/National First-Class Discipline Program of Food Science and Technology
- the BBSRC Newton Fund Joint Centre Award/the BBSRC Newton Fund Joint Centre Award
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

