Disulfide and Fully Reduced HMGB1 Induce Different Macrophage Polarization and Migration Patterns
- PMID: 34071440
- PMCID: PMC8229957
- DOI: 10.3390/biom11060800
Disulfide and Fully Reduced HMGB1 Induce Different Macrophage Polarization and Migration Patterns
Abstract
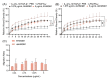
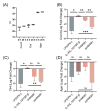
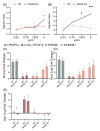
Macrophage plasticity enables cells to obtain different functions over a broad proinflammatory and repairing spectrum. In different conditions, macrophages can be induced by high-mobility group box 1 (HMGB1), a nuclear DNA-binding protein that activates innate immunity, to polarize towards a pro- (M1) or anti-inflammatory (M2) phenotype. In this study, we investigated the phenotypes of murine bone-marrow-derived macrophages (BMDMs) induced by different HMGB1 redox isoforms in depth. Our results demonstrate that disulfide HMGB1 (dsHMGB1) induces a unique macrophage phenotype that secretes pro-inflammatory cytokines, rather than inducing metabolic changes leading to nitric oxide production. Fully reduced HMGB1 (frHMGB1) did not induce macrophage polarization. The migrating function of BMDMs was measured by scratch assay after the stimulation with dsHMGB1 and frHMGB1. Both dsHMGB1 and frHMGB1 induced cell migration. We found that dsHMGB1 mediates cytokine secretion and cellular motility, mainly through toll-like receptor 4 (TLR4). Importantly, our data shows that dsHMGB1 and frHMGB1 induce distinct BMDM polarization phenotypes, and that dsHMGB1 induces a unique phenotype differing from the classical proinflammatory macrophage phenotype.
Keywords: HMGB1; RAGE; TLR4; dsHMGB1; frHMGB1; inflammation; macrophages; migration; polarization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Ferrara M., Chialli G., Ferreira L.M., Ruggieri E., Careccia G., Preti A., Piccirillo R., Bianchi M.E., Sitia G., Venereau E. Oxidation of HMGB1 Is a Dynamically Regulated Process in Physiological and Pathological Conditions. Front. Immunol. 2020;11:1122. doi: 10.3389/fimmu.2020.01122. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

