The Putative Roles and Functions of Indel, Repetition and Duplication Events in Alphavirus Non-Structural Protein 3 Hypervariable Domain (nsP3 HVD) in Evolution, Viability and Re-Emergence
- PMID: 34071712
- PMCID: PMC8228767
- DOI: 10.3390/v13061021
The Putative Roles and Functions of Indel, Repetition and Duplication Events in Alphavirus Non-Structural Protein 3 Hypervariable Domain (nsP3 HVD) in Evolution, Viability and Re-Emergence
Abstract
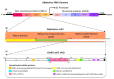
Alphavirus non-structural proteins 1-4 (nsP1, nsP2, nsP3, and nsP4) are known to be crucial for alphavirus RNA replication and translation. To date, nsP3 has been demonstrated to mediate many virus-host protein-protein interactions in several fundamental alphavirus mechanisms, particularly during the early stages of replication. However, the molecular pathways and proteins networks underlying these mechanisms remain poorly described. This is due to the low genetic sequence homology of the nsP3 protein among the alphavirus species, especially at its 3' C-terminal domain, the hypervariable domain (HVD). Moreover, the nsP3 HVD is almost or completely intrinsically disordered and has a poor ability to form secondary structures. Evolution in the nsP3 HVD region allows the alphavirus to adapt to vertebrate and insect hosts. This review focuses on the putative roles and functions of indel, repetition, and duplication events that have occurred in the alphavirus nsP3 HVD, including characterization of the differences and their implications for specificity in the context of virus-host interactions in fundamental alphavirus mechanisms, which have thus directly facilitated the evolution, adaptation, viability, and re-emergence of these viruses.
Keywords: HVD; alphavirus; duplication; emergence; evolution; indel; mutation; nsP3; phosphorylation; repetition.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

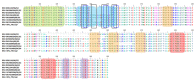

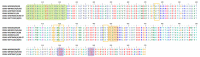
Similar articles
-
Multiple Host Factors Interact with the Hypervariable Domain of Chikungunya Virus nsP3 and Determine Viral Replication in Cell-Specific Mode.J Virol. 2018 Jul 31;92(16):e00838-18. doi: 10.1128/JVI.00838-18. Print 2018 Aug 15. J Virol. 2018. PMID: 29899097 Free PMC article.
-
Phosphorylation Sites in the Hypervariable Domain in Chikungunya Virus nsP3 Are Crucial for Viral Replication.J Virol. 2021 Apr 12;95(9):e02276-20. doi: 10.1128/JVI.02276-20. Print 2021 Apr 12. J Virol. 2021. PMID: 33568506 Free PMC article.
-
NAP1L1 and NAP1L4 Binding to Hypervariable Domain of Chikungunya Virus nsP3 Protein Is Bivalent and Requires Phosphorylation.J Virol. 2021 Jul 26;95(16):e0083621. doi: 10.1128/JVI.00836-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34076483 Free PMC article.
-
The Enigmatic Alphavirus Non-Structural Protein 3 (nsP3) Revealing Its Secrets at Last.Viruses. 2018 Feb 28;10(3):105. doi: 10.3390/v10030105. Viruses. 2018. PMID: 29495654 Free PMC article. Review.
-
Alphavirus RNA synthesis and non-structural protein functions.J Gen Virol. 2015 Sep;96(9):2483-2500. doi: 10.1099/jgv.0.000249. Epub 2015 Jul 24. J Gen Virol. 2015. PMID: 26219641 Free PMC article. Review.
Cited by
-
The Chikungunya Virus nsP3 Macro Domain Inhibits Activation of the NF-κB Pathway.Viruses. 2025 Jan 29;17(2):191. doi: 10.3390/v17020191. Viruses. 2025. PMID: 40006946 Free PMC article.
-
Antiviral activity of myricetin glycosylated compounds isolated from Marcetia taxifolia against chikungunya virus.EXCLI J. 2023 Jul 27;22:716-731. doi: 10.17179/excli2023-6242. eCollection 2023. EXCLI J. 2023. PMID: 37662709 Free PMC article.
-
A Novel Self-Amplifying mRNA with Decreased Cytotoxicity and Enhanced Protein Expression by Macrodomain Mutations.Adv Sci (Weinh). 2024 Nov;11(43):e2402936. doi: 10.1002/advs.202402936. Epub 2024 Sep 23. Adv Sci (Weinh). 2024. PMID: 39313862 Free PMC article.
-
Identification of mosquito proteins that differentially interact with alphavirus nonstructural protein 3, a determinant of vector specificity.PLoS Negl Trop Dis. 2023 Jan 25;17(1):e0011028. doi: 10.1371/journal.pntd.0011028. eCollection 2023 Jan. PLoS Negl Trop Dis. 2023. PMID: 36696390 Free PMC article.
-
Establishment of a New Real-Time Molecular Assay for the Detection of Babanki Virus in Africa.Viruses. 2024 Nov 27;16(12):1841. doi: 10.3390/v16121841. Viruses. 2024. PMID: 39772151 Free PMC article.
References
-
- Virus Pathogen Resource (ViPR) Togaviridae. [(accessed on 3 January 2020)];2020 Available online: https://www.viprbrc.org/brc/home.spg?decorator=toga.
-
- Meshram D.C., Agback P., Shiliaev N., Urakova N., Mobley J.A., Agback T., Frolova E.I., Frolov I. Multiple host factors interact with hypervariable domain of Chikungunya virus nsP3 and determine viral replication in cell-specific mode. J. Virol. 2018;92:e00838-18. doi: 10.1128/JVI.00838-18. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

