Development of a Tetraplex qPCR for the Molecular Identification and Quantification of Human Enteric Viruses, NoV and HAV, in Fish Samples
- PMID: 34071891
- PMCID: PMC8227966
- DOI: 10.3390/microorganisms9061149
Development of a Tetraplex qPCR for the Molecular Identification and Quantification of Human Enteric Viruses, NoV and HAV, in Fish Samples
Abstract
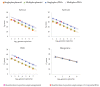
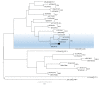
Human enteric viruses such as norovirus (NoV) and hepatitis A virus (HAV) are some of the most important causes of foodborne infections worldwide. Usually, infection via fish consumption is not a concern regarding these viruses, since fish are mainly consumed cooked. However, in the last years, raw fish consumption has become increasingly common, especially involving the use of seabass and gilthead seabream in dishes like sushi, sashimi, poke, and carpaccio. Therefore, the risk for viral infection via the consumption of raw fish has also increased. In this study, a virologic screening was performed in 323 fish specimens captured along the Portuguese coast using a tetraplex qPCR optimised for two templates (plasmid and in vitro transcribed RNA) to detect and quantify NoV GI, NoV GII and HAV genomes. A difference of approximately 1-log was found between the use of plasmid or in vitro transcribed RNA for molecular-based quantifications, showing an underestimation of genome copy-number equivalents using plasmid standard-based curves. Additionally, the presence of NoV genomic RNA in a pool of seabass brains was identified, which was shown to cluster with a major group of human norovirus sequences from genogroup I (GI.1) by phylogenetic analysis. None of the analysed fish revealed the presence of NoV GII or HAV. This result corroborates the hypothesis that enteric viruses circulate in seawater or that fish were contaminated during their transportation/handling, representing a potential risk to humans through raw or undercooked fish consumption.
Keywords: fish; hepatitis A; norovirus; pathogenic human viruses; tetraplex qPCR assay.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Bosch A., Gkogka E., Le Guyader F.S., Loisy-Hamon F., Lee A., van Lieshout L., Marthi B., Myrmel M., Sansom A., Schultz A.C., et al. Foodborne viruses: Detection, risk assessment, and control options in food processing. Int. J. Food Microbiol. 2018;285:110–128. doi: 10.1016/j.ijfoodmicro.2018.06.001. - DOI - PMC - PubMed
-
- Mangen M.J., Friesema I.H.M., Pijnacker R., Mughini L., van Pelt W. Disease Burden of Food-Related Pathogens in the Netherlands. National Institute for Public Health and the Environment; Bilthoven, The Netherlands: 2017. RIVM Lett. Rep. 2018-0037 2018. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

