The Spike of Concern-The Novel Variants of SARS-CoV-2
- PMID: 34071984
- PMCID: PMC8229995
- DOI: 10.3390/v13061002
The Spike of Concern-The Novel Variants of SARS-CoV-2
Abstract
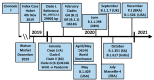
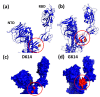
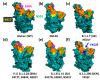
The high sequence identity of the first SARS-CoV-2 samples collected in December 2019 at Wuhan did not foretell the emergence of novel variants in the United Kingdom, North and South America, India, or South Africa that drive the current waves of the pandemic. The viral spike receptor possesses two surface areas of high mutagenic plasticity: the supersite in its N-terminal domain (NTD) that is recognised by all anti-NTD antibodies and its receptor binding domain (RBD) where 17 residues make contact with the human Ace2 protein (angiotensin I converting enzyme 2) and many neutralising antibodies bind. While NTD mutations appear at first glance very diverse, they converge on the structure of the supersite. The mutations within the RBD, on the other hand, hone in on only a small number of key sites (K417, L452, E484, N501) that are allosteric control points enabling spike to escape neutralising antibodies while maintaining or even gaining Ace2-binding activity. The D614G mutation is the hallmark of all variants, as it promotes viral spread by increasing the number of open spike protomers in the homo-trimeric receptor complex. This review discusses the recent spike mutations as well as their evolution.
Keywords: B.1.1.28; B.1.1.298; B.1.1.7; B.1.351; B.1.427; B.1.429; B.1.617; B1.526; Marseille-4; P.1; P.2.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

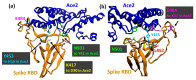

References
-
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet Lond. Engl. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

