Are Cyanobacteria an Ancestor of Chloroplasts or Just One of the Gene Donors for Plants and Algae?
- PMID: 34071987
- PMCID: PMC8227023
- DOI: 10.3390/genes12060823
Are Cyanobacteria an Ancestor of Chloroplasts or Just One of the Gene Donors for Plants and Algae?
Abstract
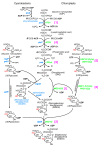
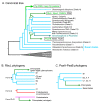
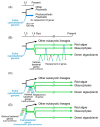
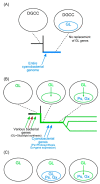
Chloroplasts of plants and algae are currently believed to originate from a cyanobacterial endosymbiont, mainly based on the shared proteins involved in the oxygenic photosynthesis and gene expression system. The phylogenetic relationship between the chloroplast and cyanobacterial genomes was important evidence for the notion that chloroplasts originated from cyanobacterial endosymbiosis. However, studies in the post-genomic era revealed that various substances (glycolipids, peptidoglycan, etc.) shared by cyanobacteria and chloroplasts are synthesized by different pathways or phylogenetically unrelated enzymes. Membranes and genomes are essential components of a cell (or an organelle), but the origins of these turned out to be different. Besides, phylogenetic trees of chloroplast-encoded genes suggest an alternative possibility that chloroplast genes could be acquired from at least three different lineages of cyanobacteria. We have to seriously examine that the chloroplast genome might be chimeric due to various independent gene flows from cyanobacteria. Chloroplast formation could be more complex than a single event of cyanobacterial endosymbiosis. I present the "host-directed chloroplast formation" hypothesis, in which the eukaryotic host cell that had acquired glycolipid synthesis genes as an adaptation to phosphate limitation facilitated chloroplast formation by providing glycolipid-based membranes (pre-adaptation). The origins of the membranes and the genome could be different, and the origin of the genome could be complex.
Keywords: Paulinella chromatophore; chloroplast origin; cyanobacterial endosymbiosis; glycolipids; host-directed chloroplast formation; peptidoglycan; phylogenetic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mereschkowsky C. Über Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Centralblatt. 1905;25:593–604.
-
- Schimper A.F.W. Über die Entwicklung der Chlorophyllkörper und Farbkörper. Bot. Z. 1883;41:105–162.
-
- Martin W., Kowallik K. Annotated English translation of Mereschkowsky’s 1905 paper ‘Über Natur und Ursprung der Chromatophoren im Pflanzenreiche’. Eur. J. Phycol. 1999;34:287–295. doi: 10.1017/S0967026299002231. - DOI
-
- Archibald J. One Plus One Equals One. Symbiosis and the Evolution of Complex Life. Oxford University Press; Oxford, UK: 2014.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

