Identification of SARS-CoV-2 Receptor Binding Inhibitors by In Vitro Screening of Drug Libraries
- PMID: 34072087
- PMCID: PMC8198929
- DOI: 10.3390/molecules26113213
Identification of SARS-CoV-2 Receptor Binding Inhibitors by In Vitro Screening of Drug Libraries
Abstract
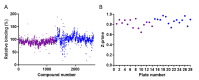
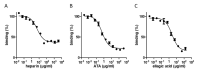
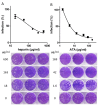
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the coronavirus disease 2019 (COVID-19) global pandemic. The first step of viral infection is cell attachment, which is mediated by the binding of the SARS-CoV-2 receptor binding domain (RBD), part of the virus spike protein, to human angiotensin-converting enzyme 2 (ACE2). Therefore, drug repurposing to discover RBD-ACE2 binding inhibitors may provide a rapid and safe approach for COVID-19 therapy. Here, we describe the development of an in vitro RBD-ACE2 binding assay and its application to identify inhibitors of the interaction of the SARS-CoV-2 RBD to ACE2 by the high-throughput screening of two compound libraries (LOPAC®1280 and DiscoveryProbeTM). Three compounds, heparin sodium, aurintricarboxylic acid (ATA), and ellagic acid, were found to exert an effective binding inhibition, with IC50 values ranging from 0.6 to 5.5 µg/mL. A plaque reduction assay in Vero E6 cells infected with a SARS-CoV-2 surrogate virus confirmed the inhibition efficacy of heparin sodium and ATA. Molecular docking analysis located potential binding sites of these compounds in the RBD. In light of these findings, the screening system described herein can be applied to other drug libraries to discover potent SARS-CoV-2 inhibitors.
Keywords: COVID-19; SARS-CoV-2; angiotensin-converting enzyme 2 (ACE2), high-throughput screening; drug repurposing; receptor binding domain (RBD); small molecule inhibitors (SMIs); spike protein.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- WHO World Health Organization. [(accessed on 8 January 2021)]; Available online: https://www.who.int/
-
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

