Detection of SARS-CoV-2 RNA by a Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Melting Curves Analysis
- PMID: 34072209
- PMCID: PMC8197939
- DOI: 10.3390/ijms22115743
Detection of SARS-CoV-2 RNA by a Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Melting Curves Analysis
Abstract
Loop-mediated isothermal amplification (LAMP) is a method of nucleic acid amplification that is more stable and resistant to DNA amplification inhibitors than conventional PCR. LAMP multiplexing with reverse transcription allows for the single-tube amplification of several RNA fragments, including an internal control sample, which provides the option of controlling all analytical steps. We developed a method of SARS-CoV-2 viral RNA detection based on multiplex reverse-transcription LAMP, with single-tube qualitative analysis of SARS-CoV-2 RNA and MS2 phage used as a control RNA. The multiplexing is based on the differences in characteristic melting peaks generated during the amplification process. The developed technique detects at least 20 copies of SARS-CoV-2 RNA per reaction on a background of 12,000 MS2 RNA copies. The total time of analysis does not exceed 40 min. The method validation, performed on 125 clinical samples of patients' nasal swabs, showed a 97.6% concordance rate with the results of real-time (RT)-PCR assays. The developed multiplexed LAMP can be employed as an alternative to PCR in diagnostic practice to save personnel and equipment time.
Keywords: LAMP; SARS-CoV-2; coronavirus; loop-mediated isothermal amplification; melting curve analysis; multiplex amplification.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

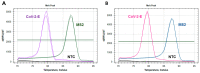
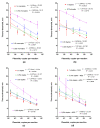
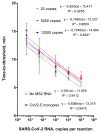
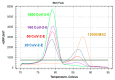
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

