Development of a New DNA Marker for Fusarium Yellows Resistance in Brassica rapa Vegetables
- PMID: 34072246
- PMCID: PMC8229042
- DOI: 10.3390/plants10061082
Development of a New DNA Marker for Fusarium Yellows Resistance in Brassica rapa Vegetables
Abstract
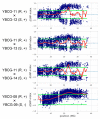
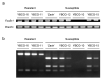
In vegetables of Brassica rapa L., Fusarium oxysporum f. sp. rapae (For) or F. oxysporum f. sp. conglutinans (Foc) cause Fusarium yellows. A resistance gene against Foc (FocBr1) has been identified, and deletion of this gene results in susceptibility (focbr1-1). In contrast, a resistance gene against For has not been identified. Inoculation tests showed that lines resistant to Foc were also resistant to For, and lines susceptible to Foc were susceptible to For. However, prediction of disease resistance by a dominant DNA marker on FocBr1 (Bra012688m) was not associated with disease resistance of For in some komatsuna lines using an inoculation test. QTL-seq using four F2 populations derived from For susceptible and resistant lines showed one causative locus on chromosome A03, which covers FocBr1. Comparison of the amino acid sequence of FocBr1 between susceptible and resistant alleles (FocBr1 and FocBo1) showed that six amino acid differences were specific to susceptible lines. The presence and absence of FocBr1 is consistent with For resistance in F2 populations. These results indicate that FocBr1 is essential for For resistance, and changed amino acid sequences result in susceptibility to For. This susceptible allele is termed focbr1-2, and a new DNA marker (focbr1-2m) for detection of the focbr1-2 allele was developed.
Keywords: Brassica rapa; DNA marker; Fusarium oxysporum f. sp. rapae; Fusarium yellows; QTL-seq; R gene; marker-assisted selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y., Liu B., Liang J., Zhuang M., Liu Y., et al. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016;48:1218–1224. doi: 10.1038/ng.3634. - DOI - PubMed
-
- Lv H., Miyaji N., Osabe K., Akter A., Mehraj H., Shea D.J., Fujimoto R. The importance of genetic and epigenetic research in the Brassica vegetables in the face of climate change. In: Kole C., editor. Genomic Designing of Climate-Smart Vegetable Crops. Springer; Berlin/Heidelberg, Germany: 2020. pp. 161–255.
-
- Mehraj H., Akter A., Miyaji N., Miyazaki J., Shea D.J., Fujimoto R., Doullah M.A.U. Genetics of clubroot and Fusarium wilt disease resistance in Brassica vegetables: The application of marker assisted breeding for disease resistance. Plants. 2020;9:726. doi: 10.3390/plants9060726. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

