Dopamine System, NMDA Receptor and EGF Family Expressions in Brain Structures of Bl6 and 129Sv Strains Displaying Different Behavioral Adaptation
- PMID: 34072341
- PMCID: PMC8227283
- DOI: 10.3390/brainsci11060725
Dopamine System, NMDA Receptor and EGF Family Expressions in Brain Structures of Bl6 and 129Sv Strains Displaying Different Behavioral Adaptation
Abstract
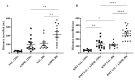
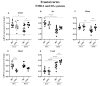
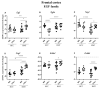
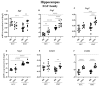
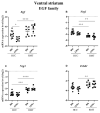
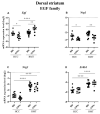
C57BL/6NTac (Bl6) and 129S6/SvEvTac (129Sv) mice display different coping strategies in stressful conditions. Our aim was to evaluate biomarkers related to different adaptation strategies in the brain of male 129Sv and Bl6 mice. We focused on signaling pathways related to the dopamine (DA) system, N-methyl-D-aspartate (NMDA) receptor and epidermal growth factor (EGF) family, shown as the key players in behavioral adaptation. Mice from Bl6 and 129Sv lines were divided into either home cage controls (HCC group) or exposed to repeated motility testing and treated with saline for 11 days (RMT group). Distinct stress responses were reflected in severe body weight loss in 129Sv and the increased exploratory behavior in Bl6 mice. Besides that, amphetamine caused significantly stronger motor stimulation in Bl6. Together with the results from gene expression (particularly Maob), this study supports higher baseline activity of DA system in Bl6. Interestingly, the adaptation is reflected with opposite changes of DA markers in dorsal and ventral striatum. In forebrain, stress increased the gene expressions of Egf-Erbb1 and Nrg1/Nrg2-Erbb4 pathways more clearly in 129Sv, whereas the corresponding proteins were significantly elevated in Bl6. We suggest that not only inhibited activity of the DA system, but also reduced activity of EGF family and NMDA receptor signaling underlies higher susceptibility to stress in 129Sv. Altogether, this study underlines the better suitability of 129Sv for modelling neuropsychiatric disorders than Bl6.
Keywords: 129Sv strain; Bl6 strain; EGF family; NMDA receptor; adaptation; dopamine system; gene expression; stress model.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

