Structural and Biophysical Characterization of the HCV E1E2 Heterodimer for Vaccine Development
- PMID: 34072451
- PMCID: PMC8227786
- DOI: 10.3390/v13061027
Structural and Biophysical Characterization of the HCV E1E2 Heterodimer for Vaccine Development
Abstract
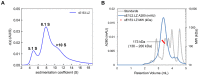
An effective vaccine for the hepatitis C virus (HCV) is a major unmet medical and public health need, and it requires an antigen that elicits immune responses to multiple key conserved epitopes. Decades of research have generated a number of vaccine candidates; based on these data and research through clinical development, a vaccine antigen based on the E1E2 glycoprotein complex appears to be the best choice. One bottleneck in the development of an E1E2-based vaccine is that the antigen is challenging to produce in large quantities and at high levels of purity and antigenic/functional integrity. This review describes the production and characterization of E1E2-based vaccine antigens, both membrane-associated and a novel secreted form of E1E2, with a particular emphasis on the major challenges facing the field and how those challenges can be addressed.
Keywords: E1; E1E2 glycoprotein complex; E2; biophysical characterization; envelope glycoproteins; hepatitis C virus (HCV); protein expression; protein purification; secreted E1E2; vaccine design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
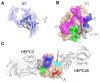
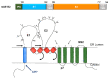



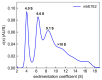
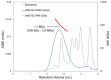
Similar articles
-
Design of a native-like secreted form of the hepatitis C virus E1E2 heterodimer.Proc Natl Acad Sci U S A. 2021 Jan 19;118(3):e2015149118. doi: 10.1073/pnas.2015149118. Proc Natl Acad Sci U S A. 2021. PMID: 33431677 Free PMC article.
-
Native Folding of a Recombinant gpE1/gpE2 Heterodimer Vaccine Antigen from a Precursor Protein Fused with Fc IgG.J Virol. 2016 Dec 16;91(1):e01552-16. doi: 10.1128/JVI.01552-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795422 Free PMC article.
-
Hepatitis C Virus E1E2 Structure, Diversity, and Implications for Vaccine Development.Viruses. 2024 May 18;16(5):803. doi: 10.3390/v16050803. Viruses. 2024. PMID: 38793684 Free PMC article. Review.
-
Fine mapping of murine antibody responses to immunization with a novel soluble form of hepatitis C virus envelope glycoprotein complex.J Virol. 2014 Sep;88(18):10459-71. doi: 10.1128/JVI.01584-14. Epub 2014 Jun 25. J Virol. 2014. PMID: 24965471 Free PMC article.
-
Computational Modeling of Hepatitis C Virus Envelope Glycoprotein Structure and Recognition.Front Immunol. 2018 May 28;9:1117. doi: 10.3389/fimmu.2018.01117. eCollection 2018. Front Immunol. 2018. PMID: 29892287 Free PMC article. Review.
Cited by
-
Where to Next? Research Directions after the First Hepatitis C Vaccine Efficacy Trial.Viruses. 2021 Jul 13;13(7):1351. doi: 10.3390/v13071351. Viruses. 2021. PMID: 34372558 Free PMC article. Review.
-
Influence of glycosylation on the immunogenicity and antigenicity of viral immunogens.Biotechnol Adv. 2024 Jan-Feb;70:108283. doi: 10.1016/j.biotechadv.2023.108283. Epub 2023 Nov 14. Biotechnol Adv. 2024. PMID: 37972669 Free PMC article. Review.
-
Virus-Mimicking Polymer Nanocomplexes Co-Assembling HCV E1E2 and Core Proteins with TLR 7/8 Agonist-Synthesis, Characterization, and In Vivo Activity.J Funct Biomater. 2025 Jan 19;16(1):34. doi: 10.3390/jfb16010034. J Funct Biomater. 2025. PMID: 39852590 Free PMC article.
-
Enhanced Production of HCV E1E2 Subunit Vaccine Candidates via Protein-Protein Interaction Identification in Glycoengineered CHO cells.bioRxiv [Preprint]. 2025 May 23:2025.05.20.655202. doi: 10.1101/2025.05.20.655202. bioRxiv. 2025. PMID: 40475571 Free PMC article. Preprint.
-
A panel of hepatitis C virus glycoproteins for the characterization of antibody responses using antibodies with diverse recognition and neutralization patterns.Virus Res. 2024 Mar;341:199308. doi: 10.1016/j.virusres.2024.199308. Epub 2024 Jan 11. Virus Res. 2024. PMID: 38171391 Free PMC article.
References
-
- WHO . Global Hepatitis Report 2017. World Health Organization; Geneva, Switzerland: 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

