PERK/ATF4-Dependent ZFAS1 Upregulation Is Associated with Sorafenib Resistance in Hepatocellular Carcinoma Cells
- PMID: 34072570
- PMCID: PMC8199104
- DOI: 10.3390/ijms22115848
PERK/ATF4-Dependent ZFAS1 Upregulation Is Associated with Sorafenib Resistance in Hepatocellular Carcinoma Cells
Abstract
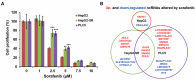
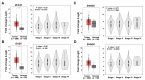
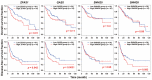
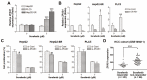
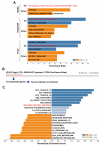
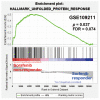
Sorafenib, a multi-kinase inhibitor, is the first-line treatment for advanced hepatocellular carcinoma (HCC) patients. However, this drug only provides a short improvement of patients' overall survival, and drug resistance is commonly developed. Thus, the identification of resistant factor(s) or biomarker(s) is needed to develop more efficient therapeutic strategies. Long, non-coding RNAs (lncRNAs) have recently been viewed as attractive cancer biomarkers and drive many important cancer phenotypes. A lncRNA, ZFAS1 (ZNFX1 antisense RNA 1) has been found to promote HCC metastasis. This study found that sorafenib induced ZFAS1 expression specifically in sorafenib-resistant HCC cells. Although ZFAS1 knockdown did not restore the sensitivity of HCC cells to sorafenib, its expression may act as a resistant biomarker for sorafenib therapy. Bioinformatics analysis predicted that sorafenib tended to induce pathways related to endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in sorafenib-resistant HCC cells. In vitro experimental evidence suggested that sorafenib induced protein kinase RNA-like ER kinase (PERK)/activating transcription factor 4 (ATF4)-dependent ZFAS1 expression, and sorafenib resistance could be overcome by PERK/ATF inhibitors. Therefore, PERK/ATF4/ZFAS1 signaling axis might be an attractive therapeutic and prognostic biomarker for sorafenib therapy in HCC.
Keywords: ER stress; drug resistance; hepatocellular carcinoma; long non-coding RNA; sorafenib; unfolded protein response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bruix J., Qin S., Merle P., Granito A., Huang Y.-H., Bodoky G., Pracht M., Yokosuka O., Rosmorduc O., Breder V., et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. - DOI - PubMed
-
- Kudo M., Finn R.S., Qin S., Han K.-H., Ikeda K., Piscaglia F., Baron A., Park J.-W., Han G., Jassem J., et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet. 2018;391:1163–1173. doi: 10.1016/S0140-6736(18)30207-1. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

