Genomic Analysis of a Novel Phage Infecting the Turkey Pathogen Escherichia coli APEC O78 and Its Endolysin Activity
- PMID: 34072620
- PMCID: PMC8229158
- DOI: 10.3390/v13061034
Genomic Analysis of a Novel Phage Infecting the Turkey Pathogen Escherichia coli APEC O78 and Its Endolysin Activity
Abstract
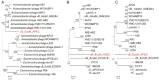
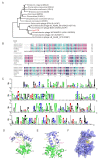
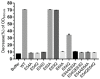
Due to the increasing spread of multidrug-resistant (MDR) bacteria, phage therapy is considered one of the most promising methods for addressing MDR bacteria. Escherichia coli lives symbiotically in the intestines of humans and some animals, and most strains are beneficial in terms of maintaining a healthy digestive tract. However, some E. coli strains can cause serious zoonotic diseases, including diarrhea, pneumonia, urinary tract infections, and hemolytic uremic syndrome. In this study, we characterized a newly isolated Myoviridae phage, vB_EcoM_APEC. The phage vB_EcoM_APEC was able to infect E. coli APEC O78, which is the most common MDR E. coli serotype in turkeys. Additionally, the phage's host range included Klebsiella pneumoniae and other E. coli strains. The genome of phage vB_EcoM_APEC (GenBank accession number MT664721) was 35,832 bp in length, with 52 putative open reading frames (ORFs) and a GC content of 41.3%. The genome of vB_EcoM_APEC exhibited low similarity (79.1% identity and 4.0% coverage) to the genome of Acinetobacter phage vB_AbaM_IME284 (GenBank no. MH853787.1) according to the nucleotide Basic Local Alignment Search Tool (BLASTn). Phylogenetic analysis revealed that vB_EcoM_APEC was a novel phage, and its genome sequence showed low similarity to other available phage genomes. Gene annotation indicated that the protein encoded by orf11 was an endolysin designated as LysO78, which exhibited 64.7% identity (91.0% coverage) with the putative endolysin of Acinetobacter baumannii phage vB_AbaM_B9. The LysO78 protein belongs to glycoside hydrolase family 19, and was described as being a chitinase class I protein. LysO78 is a helical protein with 12 α-helices containing a large domain and a small domain in terms of the predicted three-dimensional structure. The results of site-directed mutagenesis indicated that LysO78 contained the catalytic residues E54 and E64. The purified endolysin exhibited broad-spectrum bacteriolytic activity against Gram-negative strains, including the genera Klebsiella, Salmonella, Shigella, Burkholderia, Yersinia, and Pseudomonas, as well as the species Chitinimonas arctica, E. coli, Ralstonia solanacearum, and A. baumannii. An enzymatic assay showed that LysO78 had highly lytic peptidoglycan hydrolases activity (64,620,000 units/mg) against E. coli APEC O78, and that LysO78 had lytic activity in the temperature range of 4-85 °C, with an optimal temperature of 28 °C and optimal pH of 8.0, and was active at pH 3.0-12.0. Overall, the results suggested that LysO78 might be a promising therapeutic agent for controlling MDR E. coli APEC O78 and nosocomial infections caused by multidrug-resistant bacteria.
Keywords: Escherichia coli; endolysin; phage genome; turkey pathogen.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Sagi S., Konduru B., Parida M. Heterologous expression of Intimin and IpaB fusion protein in Lactococcus lactis and its mucosal delivery elicit protection against pathogenicity of Escherichia coli O157 and Shigella flexneri in a murine model. Int. Immunopharmacol. 2020;85:106617. doi: 10.1016/j.intimp.2020.106617. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

