Inferring Single-Cell 3D Chromosomal Structures Based on the Lennard-Jones Potential
- PMID: 34072879
- PMCID: PMC8199262
- DOI: 10.3390/ijms22115914
Inferring Single-Cell 3D Chromosomal Structures Based on the Lennard-Jones Potential
Abstract
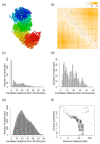
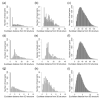
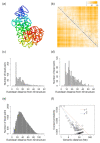
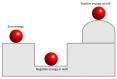
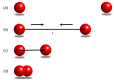
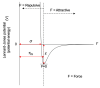
Reconstructing three-dimensional (3D) chromosomal structures based on single-cell Hi-C data is a challenging scientific problem due to the extreme sparseness of the single-cell Hi-C data. In this research, we used the Lennard-Jones potential to reconstruct both 500 kb and high-resolution 50 kb chromosomal structures based on single-cell Hi-C data. A chromosome was represented by a string of 500 kb or 50 kb DNA beads and put into a 3D cubic lattice for simulations. A 2D Gaussian function was used to impute the sparse single-cell Hi-C contact matrices. We designed a novel loss function based on the Lennard-Jones potential, in which the ε value, i.e., the well depth, was used to indicate how stable the binding of every pair of beads is. For the bead pairs that have single-cell Hi-C contacts and their neighboring bead pairs, the loss function assigns them stronger binding stability. The Metropolis-Hastings algorithm was used to try different locations for the DNA beads, and simulated annealing was used to optimize the loss function. We proved the correctness and validness of the reconstructed 3D structures by evaluating the models according to multiple criteria and comparing the models with 3D-FISH data.
Keywords: 3D chromosomal structure; 3D genome; Lennard-Jones potential; single-cell Hi-C.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

