Comparative TMT Proteomic Analysis Unveils Unique Insights into Helicoverpa armigera (Hübner) Resistance in Cajanus scarabaeoides (L.) Thouars
- PMID: 34073052
- PMCID: PMC8198728
- DOI: 10.3390/ijms22115941
Comparative TMT Proteomic Analysis Unveils Unique Insights into Helicoverpa armigera (Hübner) Resistance in Cajanus scarabaeoides (L.) Thouars
Abstract
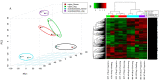
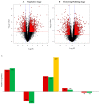
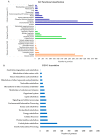
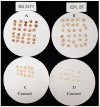
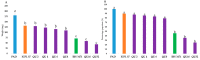
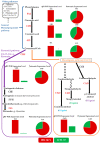
Pigeonpea [Cajanus cajan (L.) Millspaugh] is an economically important legume playing a crucial role in the semi-arid tropics. Pigeonpea is susceptible to Helicoverpa armigera (Hübner), which causes devastating yield losses. This pest is developing resistance to many commercially available insecticides. Therefore, crop wild relatives of pigeonpea, are being considered as potential sources of genes to expand the genetic base of cultivated pigeonpea to improve traits such as host plant resistance to pests and pathogens. Quantitative proteomic analysis was conducted using the tandem mass tag platform to identify differentially abundant proteins between IBS 3471 and ICPL 87 tolerant accession and susceptible variety to H. armigera, respectively. Leaf proteome were analysed at the vegetative and flowering/podding growth stages. H. armigera tolerance in IBS 3471 appeared to be related to enhanced defence responses, such as changes in secondary metabolite precursors, antioxidants, and the phenylpropanoid pathway. The development of larvae fed on an artificial diet with IBS 3471 lyophilised leaves showed similar inhibition with those fed on an artificial diet with quercetin concentrations with 32 mg/25 g of artificial diet. DAB staining (3,3'-diaminobenzidine) revealed a rapid accumulation of reactive oxygen species in IBS 3471. We conclude that IBS 3471 is an ideal candidate for improving the genetic base of cultivated pigeonpea, including traits for host plant resistance.
Keywords: Cajanus cajan; IBS 3471; ICPL 87; TMT; feeding bioassays; phenypropanoid pathway; pigeonpea; quercetin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hillocks R.J., Minja E., Mwaga A., Nahdy M.S., Subrahmanyam P. Diseases and pests of pigeonpea in eastern Africa: A review. Int. J. Pest Manag. 2000;46:7–18. doi: 10.1080/096708700227534. - DOI
-
- FAOSTAT. [(accessed on 30 September 2020)];2018 Available online: http://www.fao.org/faostat/en/
-
- Choudhary A.K., Raje R.S., Datta S., Sultana R., Ontagodi T. Conventional and Molecular Approaches towards Genetic Improvement in Pigeonpea for Insects Resistance. Am. J. Plant Sci. 2013;4:372–385. doi: 10.4236/ajps.2013.42A049. - DOI
-
- Kriticos D.J., Ota N., Hutchison W.D., Beddow J., Walsh T., Tay W.T., Borchert D.M., Paula-Moraes S.V., Czepak C., Zalucki M.P. Correction: The Potential Distribution of Invading Helicoverpa armigera in North America: Is It Just a Matter of Time? PLoS ONE. 2015;10:e0133224. doi: 10.1371/journal.pone.0133224. - DOI - PMC - PubMed
-
- Tossou E., Tepa-Yotto G., Kpindou O.K.D., Sandeu R., Datinon B., Zeukeng F., Akoton R., Tchigossou G.M., Djègbè I., Vontas J., et al. Susceptibility Profiles of Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) to Deltamethrin Reveal a Contrast between the Northern and the Southern Benin. Int. J. Environ. Res. Public Health. 2019;16:1882. doi: 10.3390/ijerph16111882. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

