Different Non-Structural Carbohydrates/Crude Proteins (NCS/CP) Ratios in Diet Shape the Gastrointestinal Microbiota of Water Buffalo
- PMID: 34073108
- PMCID: PMC8229247
- DOI: 10.3390/vetsci8060096
Different Non-Structural Carbohydrates/Crude Proteins (NCS/CP) Ratios in Diet Shape the Gastrointestinal Microbiota of Water Buffalo
Abstract
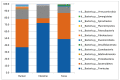
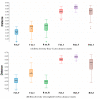
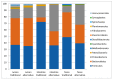
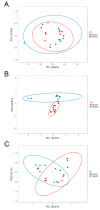
The microbiota of the gastrointestinal tract (GIT) are crucial for host health and production efficiency in ruminants. Its microbial composition can be influenced by several endogenous and exogenous factors. In the beef and dairy industry, the possibility to manipulate gut microbiota by diet and management can have important health and economic implications. The aims of this study were to characterize the different GIT site microbiota in water buffalo and evaluate the influence of diet on GIT microbiota in this animal species. We characterized and compared the microbiota of the rumen, large intestine and feces of water buffaloes fed two different diets with different non-structural carbohydrates/crude proteins (NSC/CP) ratios. Our results indicated that Bacteroidetes, Firmicutes and Proteobacteria were the most abundant phyla in all the GIT sites, with significant differences in microbiota composition between body sites both within and between groups. This result was particularly evident in the large intestine, where beta diversity analysis displayed clear clustering of samples depending on the diet. Moreover, we found a difference in diet digestibility linked to microbiota modification at the GIT level conditioned by NSC/CP levels. Diet strongly influences GIT microbiota and can therefore modulate specific GIT microorganisms able to affect the health status and performance efficiency of adult animals.
Keywords: diet; feces; fiber; food industry by-products; gastrointestinal microbiota; large intestine; rumen; tomato peel; water buffalo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Microbial Biogeography Along the Gastrointestinal Tract of a Red Panda.Front Microbiol. 2018 Jul 5;9:1411. doi: 10.3389/fmicb.2018.01411. eCollection 2018. Front Microbiol. 2018. PMID: 30026734 Free PMC article.
-
Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer.Vet Microbiol. 2013 Jun 28;164(3-4):307-14. doi: 10.1016/j.vetmic.2013.02.013. Epub 2013 Feb 24. Vet Microbiol. 2013. PMID: 23490556
-
Characterizing the microbiota in gastrointestinal tract segments of Rhabdophis subminiatus: Dynamic changes and functional predictions.Microbiologyopen. 2019 Jul;8(7):e00789. doi: 10.1002/mbo3.789. Epub 2019 Mar 7. Microbiologyopen. 2019. PMID: 30848054 Free PMC article.
-
Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes.Microorganisms. 2022 Jul 10;10(7):1391. doi: 10.3390/microorganisms10071391. Microorganisms. 2022. PMID: 35889109 Free PMC article. Review.
-
Role of diverse fermentative factors towards microbial community shift in ruminants.J Appl Microbiol. 2019 Jul;127(1):2-11. doi: 10.1111/jam.14212. Epub 2019 Mar 18. J Appl Microbiol. 2019. PMID: 30694580 Review.
Cited by
-
Comprehensive analysis of key host gene-microbe networks in the cecum tissues of the obese rabbits induced by a high-fat diet.Front Cell Infect Microbiol. 2024 Jun 14;14:1407051. doi: 10.3389/fcimb.2024.1407051. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38947127 Free PMC article.
-
Exploring the Effect of Gastrointestinal Prevotella on Growth Performance Traits in Livestock Animals.Animals (Basel). 2024 Jul 2;14(13):1965. doi: 10.3390/ani14131965. Animals (Basel). 2024. PMID: 38998077 Free PMC article. Review.
-
Comparison of Bacterial and Fungal Community Structure and Potential Function Analysis of Yak Feces before and after Weaning.Biomed Res Int. 2022 Aug 30;2022:6297231. doi: 10.1155/2022/6297231. eCollection 2022. Biomed Res Int. 2022. PMID: 36082156 Free PMC article.
References
-
- Manesco-Romagnoli E., Kmit M.C.P., Chiaramonte J.B., Rossmann M., Mendes R. Ecological Aspects on Rumen Microbiome. In: de Azevedo J.L., Quecine M.C., editors. Diversity and Benefits of Microorganisms from the Tropics. Volume 16. Springer; Cham, Switzerland: 2017. pp. 367–389. - DOI
-
- Bainbridge M.L., Cersosimo L.M., Wright A.D.G., Kraft J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein × Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS Microbiol. Ecol. 2016;92:fiw059. doi: 10.1093/femsec/fiw059. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

