Multifactorial Landscape Parses to Reveal a Predictive Model for Knee Osteoarthritis
- PMID: 34073132
- PMCID: PMC8199148
- DOI: 10.3390/ijerph18115933
Multifactorial Landscape Parses to Reveal a Predictive Model for Knee Osteoarthritis
Abstract
The present study attempted to investigate whether concerted contributions of significant risk variables, pro-inflammatory markers, and candidate genes translate into a predictive marker for knee osteoarthritis (KOA). The present study comprised 279 confirmed osteoarthritis patients (Kellgren and Lawrence scale >2) and 287 controls. Twenty SNPs within five genes (CRP, COL1A1, IL-6, VDR, and eNOS), four pro-inflammatory markers (interleukin-6 (IL-6), interleuin-1 beta (IL-1β), tumor necrosis factor alpha (TNF-α), and high sensitivity C-reactive protein (hsCRP)), along with significant risk variables were investigated. A receiver operating characteristic (ROC) curve was used to observe the predictive ability of the model for distinguishing patients with KOA. Multivariable logistic regression analysis revealed that higher body mass index (BMI), triglycerides (TG), poor sleep, IL-6, IL-1β, and hsCRP were independent predictors for KOA after adjusting for the confounding from other risk variables. Four susceptibility haplotypes for the risk of KOA, AGT, GGGGCT, AGC, and CTAAAT, were observed within CRP, IL-6, VDR, and eNOS genes, which showed their impact in recessive β(SE): 2.11 (0.76), recessive β(SE): 2.75 (0.59), dominant β(SE): 1.89 (0.52), and multiplicative modes β(SE): 1.89 (0.52), respectively. ROC curve analysis revealed the model comprising higher values of BMI, poor sleep, IL-6, and IL-1β was predictive of KOA (AUC: 0.80, 95%CI: 0.74-0.86, p< 0.001), and the strength of the predictive ability increased when susceptibility haplotypes AGC and GGGGCT were involved (AUC: 0.90, 95%CI: 0.87-0.95, p< 0.001).This study offers a predictive marker for KOA based on the risk scores of some pertinent genes and their genetic variants along with some pro-inflammatory markers and traditional risk variables.
Keywords: ROC curve analysis; genetic models; haplotypes; knee osteoarthritis; multifactorial; predictive marker.
Conflict of interest statement
None of the authors have any conflict of interest.
Figures
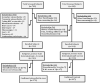
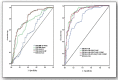
References
-
- Kwan Tat S., Padrines M., Théoleyre S., Heymann D., Fortun Y. IL-6, RANKL, TNF-alpha/IL-1: Interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev. 2004;15:49–60. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

