The Quorum Sensing Auto-Inducer 2 (AI-2) Stimulates Nitrogen Fixation and Favors Ethanol Production over Biomass Accumulation in Zymomonas mobilis
- PMID: 34073173
- PMCID: PMC8198075
- DOI: 10.3390/ijms22115628
The Quorum Sensing Auto-Inducer 2 (AI-2) Stimulates Nitrogen Fixation and Favors Ethanol Production over Biomass Accumulation in Zymomonas mobilis
Abstract
Autoinducer 2 (or AI-2) is one of the molecules used by bacteria to trigger the Quorum Sensing (QS) response, which activates expression of genes involved in a series of alternative mechanisms, when cells reach high population densities (including bioluminescence, motility, biofilm formation, stress resistance, and production of public goods, or pathogenicity factors, among others). Contrary to most autoinducers, AI-2 can induce QS responses in both Gram-negative and Gram-positive bacteria, and has been suggested to constitute a trans-specific system of bacterial communication, capable of affecting even bacteria that cannot produce this autoinducer. In this work, we demonstrate that the ethanologenic Gram-negative bacterium Zymomonas mobilis (a non-AI-2 producer) responds to exogenous AI-2 by modulating expression of genes involved in mechanisms typically associated with QS in other bacteria, such as motility, DNA repair, and nitrogen fixation. Interestingly, the metabolism of AI-2-induced Z. mobilis cells seems to favor ethanol production over biomass accumulation, probably as an adaptation to the high-energy demand of N2 fixation. This opens the possibility of employing AI-2 during the industrial production of second-generation ethanol, as a way to boost N2 fixation by these bacteria, which could reduce costs associated with the use of nitrogen-based fertilizers, without compromising ethanol production in industrial plants.
Keywords: AI-2; N2 fixation; Zymomonas mobilis; ethanol production; quorum sensing; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

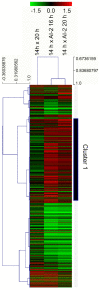
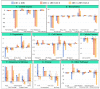
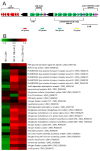
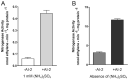

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

