Origin, Evolution and Stability of Overlapping Genes in Viruses: A Systematic Review
- PMID: 34073395
- PMCID: PMC8227390
- DOI: 10.3390/genes12060809
Origin, Evolution and Stability of Overlapping Genes in Viruses: A Systematic Review
Abstract
During their long evolutionary history viruses generated many proteins de novo by a mechanism called "overprinting". Overprinting is a process in which critical nucleotide substitutions in a pre-existing gene can induce the expression of a novel protein by translation of an alternative open reading frame (ORF). Overlapping genes represent an intriguing example of adaptive conflict, because they simultaneously encode two proteins whose freedom to change is constrained by each other. However, overlapping genes are also a source of genetic novelties, as the constraints under which alternative ORFs evolve can give rise to proteins with unusual sequence properties, most importantly the potential for novel functions. Starting with the discovery of overlapping genes in phages infecting Escherichia coli, this review covers a range of studies dealing with detection of overlapping genes in small eukaryotic viruses (genomic length below 30 kb) and recognition of their critical role in the evolution of pathogenicity. Origin of overlapping genes, what factors favor their birth and retention, and how they manage their inherent adaptive conflict are extensively reviewed. Special attention is paid to the assembly of overlapping genes into ad hoc databases, suitable for future studies, and to the development of statistical methods for exploring viral genome sequences in search of undiscovered overlaps.
Keywords: asymmetric evolution; codon usage; de novo protein creation; modular evolution; multivariate statistics; negative selection: phylogenetic distribution; positive selection; prediction methods; sequence-composition features; symmetric evolution; virus evolution.
Conflict of interest statement
The author declares no conflict of interest.
Figures



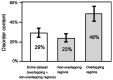
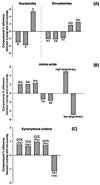
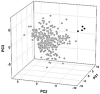
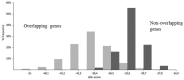
Similar articles
-
Asymmetric evolution in viral overlapping genes is a source of selective protein adaptation.Virology. 2019 Jun;532:39-47. doi: 10.1016/j.virol.2019.03.017. Epub 2019 Apr 3. Virology. 2019. PMID: 31004987 Free PMC article.
-
Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation.J Virol. 2009 Oct;83(20):10719-36. doi: 10.1128/JVI.00595-09. Epub 2009 Jul 29. J Virol. 2009. PMID: 19640978 Free PMC article.
-
Gene Birth Contributes to Structural Disorder Encoded by Overlapping Genes.Genetics. 2018 Sep;210(1):303-313. doi: 10.1534/genetics.118.301249. Epub 2018 Jul 19. Genetics. 2018. PMID: 30026186 Free PMC article.
-
[The great virus comeback].Biol Aujourdhui. 2013;207(3):153-68. doi: 10.1051/jbio/2013018. Epub 2013 Dec 13. Biol Aujourdhui. 2013. PMID: 24330969 Review. French.
-
Molecular evolution in papova viruses and in bacteriophages.Adv Biophys. 1982;15:1-17. doi: 10.1016/0065-227x(82)90003-x. Adv Biophys. 1982. PMID: 6285681 Review.
Cited by
-
Hepatitis B and circadian rhythm of the liver.World J Gastroenterol. 2022 Jul 21;28(27):3282-3296. doi: 10.3748/wjg.v28.i27.3282. World J Gastroenterol. 2022. PMID: 36158265 Free PMC article. Review.
-
Structural conservation of HBV-like capsid proteins over hundreds of millions of years despite the shift from non-enveloped to enveloped life-style.Nat Commun. 2023 Mar 22;14(1):1574. doi: 10.1038/s41467-023-37068-w. Nat Commun. 2023. PMID: 36949039 Free PMC article.
-
Variable Proportions of Phylogenetic Clustering and Low Levels of Antiviral Drug Resistance among the Major HBV Sub-Genotypes in the Middle East and North Africa.Pathogens. 2021 Oct 15;10(10):1333. doi: 10.3390/pathogens10101333. Pathogens. 2021. PMID: 34684283 Free PMC article.
-
Are There Hidden Genes in DNA/RNA Vaccines?Front Immunol. 2022 Feb 8;13:801915. doi: 10.3389/fimmu.2022.801915. eCollection 2022. Front Immunol. 2022. PMID: 35211117 Free PMC article.
-
Endemism shapes viral ecology and evolution in globally distributed hydrothermal vent ecosystems.Nat Commun. 2025 May 1;16(1):4076. doi: 10.1038/s41467-025-59154-x. Nat Commun. 2025. PMID: 40307239 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

