Biomedical Text Link Prediction for Drug Discovery: A Case Study with COVID-19
- PMID: 34073456
- PMCID: PMC8230210
- DOI: 10.3390/pharmaceutics13060794
Biomedical Text Link Prediction for Drug Discovery: A Case Study with COVID-19
Abstract
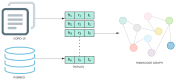
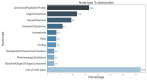
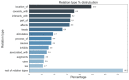
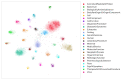
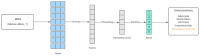
Link prediction in artificial intelligence is used to identify missing links or derive future relationships that can occur in complex networks. A link prediction model was developed using the complex heterogeneous biomedical knowledge graph, SemNet, to predict missing links in biomedical literature for drug discovery. A web application visualized knowledge graph embeddings and link prediction results using TransE, CompleX, and RotatE based methods. The link prediction model achieved up to 0.44 hits@10 on the entity prediction tasks. The recent outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as COVID-19, served as a case study to demonstrate the efficacy of link prediction modeling for drug discovery. The link prediction algorithm guided identification and ranking of repurposed drug candidates for SARS-CoV-2 primarily by text mining biomedical literature from previous coronaviruses, including SARS and middle east respiratory syndrome (MERS). Repurposed drugs included potential primary SARS-CoV-2 treatment, adjunctive therapies, or therapeutics to treat side effects. The link prediction accuracy for nodes ranked highly for SARS coronavirus was 0.875 as calculated by human in the loop validation on existing COVID-19 specific data sets. Drug classes predicted as highly ranked include anti-inflammatory, nucleoside analogs, protease inhibitors, antimalarials, envelope proteins, and glycoproteins. Examples of highly ranked predicted links to SARS-CoV-2: human leukocyte interferon, recombinant interferon-gamma, cyclosporine, antiviral therapy, zidovudine, chloroquine, vaccination, methotrexate, artemisinin, alkaloids, glycyrrhizic acid, quinine, flavonoids, amprenavir, suramin, complement system proteins, fluoroquinolones, bone marrow transplantation, albuterol, ciprofloxacin, quinolone antibacterial agents, and hydroxymethylglutaryl-CoA reductase inhibitors. Approximately 40% of identified drugs were not previously connected to SARS, such as edetic acid or biotin. In summary, link prediction can effectively suggest repurposed drugs for emergent diseases.
Keywords: COVID-19; SARS-CoV-2; coronavirus; literature review; machine learning; natural language processing; repurposed drugs; text mining.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



Similar articles
-
Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning.Comput Struct Biotechnol J. 2021;19:3133-3148. doi: 10.1016/j.csbj.2021.05.037. Epub 2021 May 24. Comput Struct Biotechnol J. 2021. PMID: 34055238 Free PMC article.
-
Drug repurposing for COVID-19 via knowledge graph completion.J Biomed Inform. 2021 Mar;115:103696. doi: 10.1016/j.jbi.2021.103696. Epub 2021 Feb 8. J Biomed Inform. 2021. PMID: 33571675 Free PMC article.
-
Potential Target Discovery and Drug Repurposing for Coronaviruses: Study Involving a Knowledge Graph-Based Approach.J Med Internet Res. 2023 Oct 20;25:e45225. doi: 10.2196/45225. J Med Internet Res. 2023. PMID: 37862061 Free PMC article.
-
Role of biological Data Mining and Machine Learning Techniques in Detecting and Diagnosing the Novel Coronavirus (COVID-19): A Systematic Review.J Med Syst. 2020 May 25;44(7):122. doi: 10.1007/s10916-020-01582-x. J Med Syst. 2020. PMID: 32451808 Free PMC article.
-
Human and novel coronavirus infections in children: a review.Paediatr Int Child Health. 2021 Feb;41(1):36-55. doi: 10.1080/20469047.2020.1781356. Epub 2020 Jun 25. Paediatr Int Child Health. 2021. PMID: 32584199 Review.
Cited by
-
Paving New Roads Using Allium sativum as a Repurposed Drug and Analyzing its Antiviral Action Using Artificial Intelligence Technology.Iran J Pharm Res. 2023 Jan 21;21(1):e131577. doi: 10.5812/ijpr-131577. eCollection 2022 Dec. Iran J Pharm Res. 2023. PMID: 36915406 Free PMC article. Review.
-
Optimizations for Computing Relatedness in Biomedical Heterogeneous Information Networks: SemNet 2.0.Big Data Cogn Comput. 2022 Mar;6(1):27. doi: 10.3390/bdcc6010027. Epub 2022 Mar 1. Big Data Cogn Comput. 2022. PMID: 35936510 Free PMC article.
-
Data-Driven Technology Roadmaps to Identify Potential Technology Opportunities for Hyperuricemia Drugs.Pharmaceuticals (Basel). 2022 Nov 3;15(11):1357. doi: 10.3390/ph15111357. Pharmaceuticals (Basel). 2022. PMID: 36355529 Free PMC article.
-
A Systematic Review on the Contribution of Artificial Intelligence in the Development of Medicines for COVID-2019.J Pers Med. 2021 Sep 18;11(9):926. doi: 10.3390/jpm11090926. J Pers Med. 2021. PMID: 34575703 Free PMC article. Review.
-
Literature-Based Discovery to Elucidate the Biological Links between Resistant Hypertension and COVID-19.Biology (Basel). 2023 Sep 21;12(9):1269. doi: 10.3390/biology12091269. Biology (Basel). 2023. PMID: 37759668 Free PMC article.
References
-
- Wang L.L., Lo K., Chandrasekhar Y., Reas R., Yang J., Eide D., Funk K., Kinney R., Liu Z., Merrill W., et al. CORD-19: The Covid-19 Open Research Dataset. arXiv. 20202004.10706
-
- Wilcke X., Bloem P., De Boer V. The knowledge graph as the default data model for learning on heterogeneous knowledge. Data Sci. 2017;1:39–57. doi: 10.3233/DS-170007. - DOI
-
- Bordes A., Usunier N., Garcia-Duran A., Weston J., Yakhnenko O. Translating embeddings for modeling multi-relational data; Proceedings of the Advances in Neural Information Processing Systems; Lake Tahoe, NV, USA. 5–8 December 2013; pp. 2787–2795.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

