Presence and Diversity of Different Enteric Viruses in Wild Norway Rats (Rattus norvegicus)
- PMID: 34073462
- PMCID: PMC8227696
- DOI: 10.3390/v13060992
Presence and Diversity of Different Enteric Viruses in Wild Norway Rats (Rattus norvegicus)
Abstract
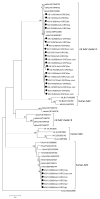
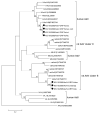
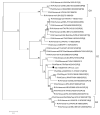
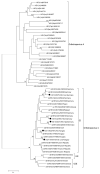
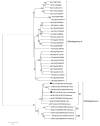
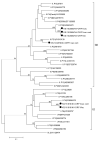
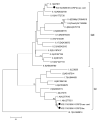
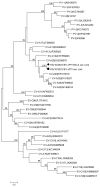
Rodents are common reservoirs for numerous zoonotic pathogens, but knowledge about diversity of pathogens in rodents is still limited. Here, we investigated the occurrence and genetic diversity of enteric viruses in 51 Norway rats collected in three different countries in Europe. RNA of at least one virus was detected in the intestine of 49 of 51 animals. Astrovirus RNA was detected in 46 animals, mostly of rat astroviruses. Human astrovirus (HAstV-8) RNA was detected in one, rotavirus group A (RVA) RNA was identified in eleven animals. One RVA RNA could be typed as rat G3 type. Rat hepatitis E virus (HEV) RNA was detected in five animals. Two entire genome sequences of ratHEV were determined. Human norovirus RNA was detected in four animals with the genotypes GI.P4-GI.4, GII.P33-GII.1, and GII.P21. In one animal, a replication competent coxsackievirus A20 strain was detected. Additionally, RNA of an enterovirus species A strain was detected in the same animal, albeit in a different tissue. The results show a high detection rate and diversity of enteric viruses in Norway rats in Europe and indicate their significance as vectors for zoonotic transmission of enteric viruses. The detailed role of Norway rats and transmission pathways of enteric viruses needs to be investigated in further studies.
Keywords: Norway rat; astrovirus; enterovirus; hepatitis E virus; norovirus; rodent; rotavirus; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
Similar articles
-
Exploring the Potential of Muridae as Sentinels for Human and Zoonotic Viruses.Viruses. 2024 Jun 27;16(7):1041. doi: 10.3390/v16071041. Viruses. 2024. PMID: 39066204 Free PMC article.
-
Changing pattern of prevalence, genetic diversity, and mixed infections of viruses associated with acute gastroenteritis in pediatric patients in New Delhi, India.J Med Virol. 2018 Mar;90(3):469-476. doi: 10.1002/jmv.24980. Epub 2017 Nov 17. J Med Virol. 2018. PMID: 29064572
-
Genetic Diversity of Enteric Viruses in Children under Five Years Old in Gabon.Viruses. 2021 Mar 24;13(4):545. doi: 10.3390/v13040545. Viruses. 2021. PMID: 33805214 Free PMC article.
-
Feline Virome-A Review of Novel Enteric Viruses Detected in Cats.Viruses. 2019 Sep 30;11(10):908. doi: 10.3390/v11100908. Viruses. 2019. PMID: 31575055 Free PMC article. Review.
-
Astrovirus infections in humans and animals - molecular biology, genetic diversity, and interspecies transmissions.Infect Genet Evol. 2011 Oct;11(7):1529-44. doi: 10.1016/j.meegid.2011.07.024. Epub 2011 Aug 5. Infect Genet Evol. 2011. PMID: 21843659 Free PMC article. Review.
Cited by
-
Exploring the Potential of Muridae as Sentinels for Human and Zoonotic Viruses.Viruses. 2024 Jun 27;16(7):1041. doi: 10.3390/v16071041. Viruses. 2024. PMID: 39066204 Free PMC article.
-
Diversity of human astroviruses in Germany 2018 and 2019.Virol J. 2022 Dec 21;19(1):221. doi: 10.1186/s12985-022-01955-3. Virol J. 2022. PMID: 36544187 Free PMC article.
-
Lack of molecular evidence of fecal-borne viruses in capybaras from São Paulo state, Brazil, 2018-2020: a minor public health issue.Braz J Microbiol. 2023 Mar;54(1):543-551. doi: 10.1007/s42770-022-00859-2. Epub 2022 Nov 7. Braz J Microbiol. 2023. PMID: 36342660 Free PMC article.
-
Investigating the Hepatitis E Virus (HEV) Diversity in Rat Reservoirs from Northern Italy.Pathogens. 2024 Jul 29;13(8):633. doi: 10.3390/pathogens13080633. Pathogens. 2024. PMID: 39204234 Free PMC article.
-
Genetic and biological characteristics of species A rotaviruses detected in common shrews suggest a distinct evolutionary trajectory.Virus Evol. 2022 Jan 28;8(1):veac004. doi: 10.1093/ve/veac004. eCollection 2022. Virus Evol. 2022. PMID: 35169491 Free PMC article.
References
-
- Wilson D.E., Reeder D.M. Mammal Species of the World: A Taxonomic and Geographic Reference. 3rd ed. Johns Hopkins University Press; Baltimore, ML, USA: 2005.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

