Genome Evolutionary Dynamics Meets Functional Genomics: A Case Story on the Identification of SLC25A44
- PMID: 34073512
- PMCID: PMC8199184
- DOI: 10.3390/ijms22115669
Genome Evolutionary Dynamics Meets Functional Genomics: A Case Story on the Identification of SLC25A44
Abstract
Gene clusters are becoming promising tools for gene identification. The study reveals the purposive genomic distribution of genes toward higher inheritance rates of intact metabolic pathways/phenotypes and, thereby, higher fitness. The co-localization of co-expressed, co-interacting, and functionally related genes was found as genome-wide trends in humans, mouse, golden eagle, rice fish, Drosophila, peanut, and Arabidopsis. As anticipated, the analyses verified the co-segregation of co-localized events. A negative correlation was notable between the likelihood of co-localization events and the inter-loci distances. The evolution of genomic blocks was also found convergent and uniform along the chromosomal arms. Calling a genomic block responsible for adjacent metabolic reactions is therefore recommended for identification of candidate genes and interpretation of cellular functions. As a case story, a function in the metabolism of energy and secondary metabolites was proposed for Slc25A44, based on its genomic local information. Slc25A44 was further characterized as an essential housekeeping gene which has been under evolutionary purifying pressure and belongs to the phylogenetic ETC-clade of SLC25s. Pathway enrichment mapped the Slc25A44s to the energy metabolism. The expression of peanut and human Slc25A44s in oocytes and Saccharomyces cerevisiae strains confirmed the transport of common precursors for secondary metabolites and ubiquinone. These results suggest that SLC25A44 is a mitochondrion-ER-nucleus zone transporter with biotechnological applications. Finally, a conserved three-amino acid signature on the cytosolic face of transport cavity was found important for rational engineering of SLC25s.
Keywords: SLC25 subfamily 44; candidate gene; electron transfer chains; flavonoids; gene clusters; gene organization; genomic co-localization; para-coumaric acid; resveratrol; ubiquinone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

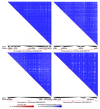





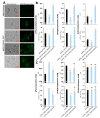

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

