Could Histamine H1 Receptor Antagonists Be Used for Treating COVID-19?
- PMID: 34073529
- PMCID: PMC8199351
- DOI: 10.3390/ijms22115672
Could Histamine H1 Receptor Antagonists Be Used for Treating COVID-19?
Abstract
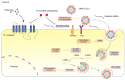
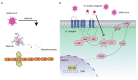
COVID-19 has rapidly become a pandemic worldwide, causing extensive and long-term health issues. There is an urgent need to identify therapies that limit SARS-CoV-2 infection and improve the outcome of COVID-19 patients. Unbalanced lung inflammation is a common feature in severe COVID-19 patients; therefore, reducing lung inflammation can undoubtedly benefit the clinical manifestations. Histamine H1 receptor (H1 receptor) antagonists are widely prescribed medications to treat allergic diseases, while recently it has emerged that they show significant promise as anti-SARS-CoV-2 agents. Here, we briefly summarize the novel use of H1 receptor antagonists in combating SARS-CoV-2 infection. We also describe the potential antiviral mechanisms of H1 receptor antagonists on SARS-CoV-2. Finally, the opportunities and challenges of the use of H1 receptor antagonists in managing COVID-19 are discussed.
Keywords: COVID-19; H1 receptor antagonists; NF-κB signaling; drugs; treatment.
Conflict of interest statement
All the authors declare that they have no conflict of interest.
Figures
Similar articles
-
Repositioning of histamine H1 receptor antagonist: Doxepin inhibits viropexis of SARS-CoV-2 Spike pseudovirus by blocking ACE2.Eur J Pharmacol. 2021 Apr 5;896:173897. doi: 10.1016/j.ejphar.2021.173897. Epub 2021 Jan 23. Eur J Pharmacol. 2021. PMID: 33497607 Free PMC article.
-
Histamine H1-receptor antagonists inhibit nuclear factor-kappaB and activator protein-1 activities via H1-receptor-dependent and -independent mechanisms.Clin Exp Allergy. 2008 Jun;38(6):947-56. doi: 10.1111/j.1365-2222.2008.02990.x. Clin Exp Allergy. 2008. PMID: 18498541
-
Phillyrin (KD-1) exerts anti-viral and anti-inflammatory activities against novel coronavirus (SARS-CoV-2) and human coronavirus 229E (HCoV-229E) by suppressing the nuclear factor kappa B (NF-κB) signaling pathway.Phytomedicine. 2020 Nov;78:153296. doi: 10.1016/j.phymed.2020.153296. Epub 2020 Aug 1. Phytomedicine. 2020. PMID: 32890913 Free PMC article.
-
Histamine receptors and COVID-19.Inflamm Res. 2021 Jan;70(1):67-75. doi: 10.1007/s00011-020-01422-1. Epub 2020 Nov 18. Inflamm Res. 2021. PMID: 33206207 Free PMC article.
-
The Role of the Nuclear Factor-Kappa B (NF-κB) Pathway in SARS-CoV-2 Infection.Pathogens. 2024 Feb 12;13(2):164. doi: 10.3390/pathogens13020164. Pathogens. 2024. PMID: 38392902 Free PMC article. Review.
Cited by
-
Covid-19 Histamine theory: Why antihistamines should be incorporated as the basic component in Covid-19 management?Health Sci Rep. 2023 Feb 7;6(2):e1109. doi: 10.1002/hsr2.1109. eCollection 2023 Feb. Health Sci Rep. 2023. PMID: 36778771 Free PMC article. No abstract available.
-
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) and Comorbidities: Linked by Vascular Pathomechanisms and Vasoactive Mediators?Medicina (Kaunas). 2023 May 18;59(5):978. doi: 10.3390/medicina59050978. Medicina (Kaunas). 2023. PMID: 37241210 Free PMC article.
-
Human Identical Sequences, hyaluronan, and hymecromone ─ the new mechanism and management of COVID-19.Mol Biomed. 2022 May 20;3(1):15. doi: 10.1186/s43556-022-00077-0. Mol Biomed. 2022. PMID: 35593963 Free PMC article. Review.
-
Carbon dioxide and MAPK signalling: towards therapy for inflammation.Cell Commun Signal. 2023 Oct 10;21(1):280. doi: 10.1186/s12964-023-01306-x. Cell Commun Signal. 2023. PMID: 37817178 Free PMC article. Review.
-
Pathological mechanisms of glial cell activation and neurodegenerative and neuropsychiatric disorders caused by Toxoplasma gondii infection.Front Microbiol. 2024 Dec 11;15:1512233. doi: 10.3389/fmicb.2024.1512233. eCollection 2024. Front Microbiol. 2024. PMID: 39723133 Free PMC article. Review.
References
-
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

