Learning from Embryogenesis-A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development
- PMID: 34073664
- PMCID: PMC8197100
- DOI: 10.3390/jcm10112259
Learning from Embryogenesis-A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development
Abstract
Malignant melanoma is one of the most dangerous tumor types due to its high metastasis rates and a steadily increasing incidence. During tumorigenesis, the molecular processes of embryonic development, exemplified by epithelial-mesenchymal transition (EMT), are often reactivated. For melanoma development, the exact molecular differences between melanoblasts, melanocytes, and melanoma cells are not completely understood. In this study, we aimed to identify microRNAs (miRNAs) that promote melanoma tumorigenesis and progression, based on an in vitro model of normal human epidermal melanocyte (NHEM) de-differentiation into melanoblast-like cells (MBrCs). Using miRNA-sequencing and differential expression analysis, we demonstrated in this study that a majority of miRNAs have an almost equal expression level in NHEMs and MBrCs but are significantly differentially regulated in primary tumor- and metastasis-derived melanoma cell lines. Further, a target gene analysis of strongly regulated but functionally unknown miRNAs yielded the implication of those miRNAs in many important cellular pathways driving malignancy. We hypothesize that many of the miRNAs discovered in our study are key drivers of melanoma development as they account for the tumorigenic potential that differentiates melanoma cells from proliferating or migrating embryonic cells.
Keywords: embryogenesis; melanoblasts; melanoma; miRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


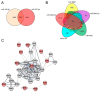
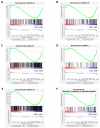
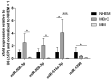
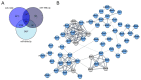
References
-
- Leiter U., Eigentler T., Garbe C. Sunlight, Vitamin D and Skin Cancer. Springer; Berlin/Heidelberg, Germany: 2014. Epidemiology of skin cancer; pp. 120–140. - PubMed
-
- Garbe C., Peris K., Hauschild A., Saiag P., Middleton M., Bastholt L., Grob J.-J., Malvehy J., Newton-Bishop J., Stratigos A.J. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline—Update 2016. Eur. J. Cancer. 2016;63:201–217. doi: 10.1016/j.ejca.2016.05.005. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

