Classification of Monocytes, Promonocytes and Monoblasts Using Deep Neural Network Models: An Area of Unmet Need in Diagnostic Hematopathology
- PMID: 34073699
- PMCID: PMC8197234
- DOI: 10.3390/jcm10112264
Classification of Monocytes, Promonocytes and Monoblasts Using Deep Neural Network Models: An Area of Unmet Need in Diagnostic Hematopathology
Abstract
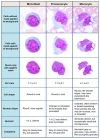
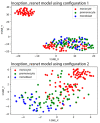
The accurate diagnosis of chronic myelomonocytic leukemia (CMML) and acute myeloid leukemia (AML) subtypes with monocytic differentiation relies on the proper identification and quantitation of blast cells and blast-equivalent cells, including promonocytes. This distinction can be quite challenging given the cytomorphologic and immunophenotypic similarities among the monocytic cell precursors. The aim of this study was to assess the performance of convolutional neural networks (CNN) in separating monocytes from their precursors (i.e., promonocytes and monoblasts). We collected digital images of 935 monocytic cells that were blindly reviewed by five experienced morphologists and assigned into three subtypes: monocyte, promonocyte, and blast. The consensus between reviewers was considered as a ground truth reference label for each cell. In order to assess the performance of CNN models, we divided our data into training (70%), validation (10%), and test (20%) datasets, as well as applied fivefold cross validation. The CNN models did not perform well for predicting three monocytic subtypes, but their performance was significantly improved for two subtypes (monocyte vs. promonocytes + blasts). Our findings (1) support the concept that morphologic distinction between monocytic cells of various differentiation level is difficult; (2) suggest that combining blasts and promonocytes into a single category is desirable for improved accuracy; and (3) show that CNN models can reach accuracy comparable to human reviewers (0.78 ± 0.10 vs. 0.86 ± 0.05). As far as we know, this is the first study to separate monocytes from their precursors using CNN.
Keywords: artificial intelligence; chronic myelomonocytic leukemia (CMML) and acute myeloid leukemia (AML) for acute monoblastic leukemia and acute monocytic leukemia; concordance between hematopathologists; digital imaging; improving diagnosis accuracy; monocytes; promonocytes and monoblasts.
Conflict of interest statement
Mohamed Salama serves on the Board of Directors and has stock option at Techcyte Inc.
Figures
Similar articles
-
Concordance among hematopathologists in classifying blasts plus promonocytes: A bone marrow pathology group study.Int J Lab Hematol. 2020 Aug;42(4):418-422. doi: 10.1111/ijlh.13212. Epub 2020 Apr 16. Int J Lab Hematol. 2020. PMID: 32297416
-
Flow cytometric analysis of different CD14 epitopes can help identify immature monocytic populations.Am J Clin Pathol. 2005 Dec;124(6):930-6. Am J Clin Pathol. 2005. PMID: 16416743
-
Characteristics and functions of monocytes and promonocytes in monocytic leukemia.Haematol Blood Transfus. 1981;27:175-8. doi: 10.1007/978-3-642-81696-3_19. Haematol Blood Transfus. 1981. PMID: 6948756
-
Diagnostic challenges in acute monoblastic/monocytic leukemia in children.Front Pediatr. 2022 Sep 28;10:911093. doi: 10.3389/fped.2022.911093. eCollection 2022. Front Pediatr. 2022. PMID: 36245718 Free PMC article. Review.
-
Acute monoblastic leukemia with t(8;16): a distinct clinicopathologic entity; report of a case and review of the literature.Am J Hematol. 2001 Mar;66(3):207-12. doi: 10.1002/1096-8652(200103)66:3<207::aid-ajh1046>3.0.co;2-q. Am J Hematol. 2001. PMID: 11279628 Review.
Cited by
-
Clinical Applications of Artificial Intelligence-An Updated Overview.J Clin Med. 2022 Apr 18;11(8):2265. doi: 10.3390/jcm11082265. J Clin Med. 2022. PMID: 35456357 Free PMC article. Review.
-
DeepHeme: A generalizable, bone marrow classifier with hematopathologist-level performance.bioRxiv [Preprint]. 2023 Feb 21:2023.02.20.528987. doi: 10.1101/2023.02.20.528987. bioRxiv. 2023. PMID: 36865216 Free PMC article. Preprint.
-
Short histological kaleidoscope - recent findings in histology. Part III.Rom J Morphol Embryol. 2023 Apr-Jun;64(2):115-133. doi: 10.47162/RJME.64.2.01. Rom J Morphol Embryol. 2023. PMID: 37518868 Free PMC article.
-
Evaluation of the practical application of the category-imbalanced myeloid cell classification model.PLoS One. 2025 Jan 30;20(1):e0313277. doi: 10.1371/journal.pone.0313277. eCollection 2025. PLoS One. 2025. PMID: 39883644 Free PMC article.
-
An integrative multiparametric approach stratifies putative distinct phenotypes of blast phase chronic myelomonocytic leukemia.Cell Rep Med. 2025 Feb 18;6(2):101933. doi: 10.1016/j.xcrm.2025.101933. Epub 2025 Jan 31. Cell Rep Med. 2025. PMID: 39892394 Free PMC article.
References
-
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. - DOI - PubMed
-
- Campo E., Harris N.L., Pileri S.A., Jaffe E.S., Stein H., Thiele J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. IARC Who Classification of Tum; Lyon, France: 2017.
-
- Arber D.A. Acute myeloid leukaemia, not otherwise specified. In: Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., editors. World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues. Revised 4th ed. IARC Press; Lyon, France: 2017. pp. 156–166.
-
- Bain B., Bain B.J., Matutes E. Chronic Myeloid Leukaemias. Clinical Publishing, Atlas Medical Pub Ltd.; New York, NY, USA: 2012.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

