Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection
- PMID: 34073756
- PMCID: PMC8225209
- DOI: 10.3390/bios11060167
Clinical Utility of Biosensing Platforms for Confirmation of SARS-CoV-2 Infection
Abstract
Despite collaborative efforts from all countries, coronavirus disease 2019 (COVID-19) pandemic has been continuing to spread globally, forcing the world into social distancing period, making a special challenge for public healthcare system. Before vaccine widely available, the best approach to manage severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is to achieve highest diagnostic accuracy by improving biosensor efficacy. For SARS-CoV-2 diagnostics, intensive attempts have been made by many scientists to ameliorate the drawback of current biosensors of SARS-CoV-2 in clinical diagnosis to offer benefits related to platform proposal, systematic analytical methods, system combination, and miniaturization. This review assesses ongoing research efforts aimed at developing integrated diagnostic tools to detect RNA viruses and their biomarkers for clinical diagnostics of SARS-CoV-2 infection and further highlights promising technology for SARS-CoV-2 specific diagnosis. The comparisons of SARS-CoV-2 biomarkers as well as their applicable biosensors in the field of clinical diagnosis were summarized to give scientists an advantage to develop superior diagnostic platforms. Furthermore, this review describes the prospects for this rapidly growing field of diagnostic research, raising further interest in analytical technology and strategic plan for future pandemics.
Keywords: COVID-19 clinical diagnostics; ELISA; RT-LAMP; RT-PCR; SARS-CoV-2; electrochemical biosensor; lab-in-a-tube; lateral flow assay; nucleic acid amplification; optical biosensor.
Conflict of interest statement
There is no known competing financial interest.
Figures
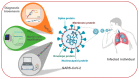




Similar articles
-
Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen.Mikrochim Acta. 2021 May 26;188(6):199. doi: 10.1007/s00604-021-04867-1. Mikrochim Acta. 2021. PMID: 34041585 Free PMC article.
-
The potential application of electrochemical biosensors in the COVID-19 pandemic: A perspective on the rapid diagnostics of SARS-CoV-2.Biosens Bioelectron. 2021 Mar 15;176:112905. doi: 10.1016/j.bios.2020.112905. Epub 2020 Dec 17. Biosens Bioelectron. 2021. PMID: 33358285 Free PMC article. Review.
-
Rapid SARS-CoV-2 Detection Using Electrochemical Immunosensor.Sensors (Basel). 2021 Jan 8;21(2):390. doi: 10.3390/s21020390. Sensors (Basel). 2021. PMID: 33429915 Free PMC article.
-
iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2.Virus Res. 2020 Oct 15;288:198129. doi: 10.1016/j.virusres.2020.198129. Epub 2020 Aug 18. Virus Res. 2020. PMID: 32822689 Free PMC article.
-
Rapid diagnosis of SARS-CoV-2 using potential point-of-care electrochemical immunosensor: Toward the future prospects.Int Rev Immunol. 2021;40(1-2):126-142. doi: 10.1080/08830185.2021.1872566. Epub 2021 Jan 15. Int Rev Immunol. 2021. PMID: 33448909 Review.
Cited by
-
Rational Programming of Cas12a for Early-Stage Detection of COVID-19 by Lateral Flow Assay and Portable Real-Time Fluorescence Readout Facilities.Biosensors (Basel). 2021 Dec 26;12(1):11. doi: 10.3390/bios12010011. Biosensors (Basel). 2021. PMID: 35049639 Free PMC article.
-
Using Nanomaterials for SARS-CoV-2 Sensing via Electrochemical Techniques.Micromachines (Basel). 2023 Apr 25;14(5):933. doi: 10.3390/mi14050933. Micromachines (Basel). 2023. PMID: 37241556 Free PMC article. Review.
References
-
- Maganga G.D., Pinto A., Mombo I.M., Madjitobaye M., Beyeme A.M.M., Boundenga L., Gouilh M.A., N’Dilimabaka N., Drexler J.F., Drosten C., et al. Genetic diversity and ecology of coronaviruses hosted by cave-dwelling bats in Gabon. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-64159-1. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

