Comparative Analysis of Mitochondrial Genome Features among Four Clonostachys Species and Insight into Their Systematic Positions in the Order Hypocreales
- PMID: 34073831
- PMCID: PMC8197242
- DOI: 10.3390/ijms22115530
Comparative Analysis of Mitochondrial Genome Features among Four Clonostachys Species and Insight into Their Systematic Positions in the Order Hypocreales
Abstract
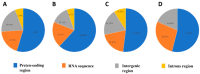
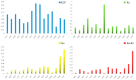
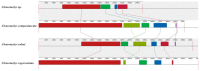
The mycoparasite fungi of Clonostachys have contributed to the biological control of plant fungal disease and nematodes. The Clonostachys fungi strains were isolated from Ophiocordyceps highlandensis, Ophiocordycepsnigrolla and soil, which identified as Clonostachyscompactiuscula, Clonostachysrogersoniana, Clonostachyssolani and Clonostachys sp. To explore the evolutionary relationship between the mentioned species, the mitochondrial genomes of four Clonostachys species were sequenced and assembled. The four mitogenomes consisted of complete circular DNA molecules, with the total sizes ranging from 27,410 bp to 42,075 bp. The GC contents, GC skews and AT skews of the mitogenomes varied considerably. Mitogenomic synteny analysis indicated that these mitogenomes underwent gene rearrangements. Among the 15 protein-coding genes within the mitogenomes, the nad4L gene exhibited the least genetic distance, demonstrating a high degree of conservation. The selection pressure analysis of these 15 PCGs were all below 1, indicating that PCGs were subject to purifying selection. Based on protein-coding gene calculation of the significantly supported topologies, the four Clonostachys species were divided into a group in the phylogenetic tree. The results supplemented the database of mitogenomes in Hypocreales order, which might be a useful research tool to conduct a phylogenetic analysis of Clonostachys. Additionally, the suitable molecular marker was significant to study phylogenetic relationships in the Bionectriaceae family.
Keywords: Clonostachys; mitochondrial genome; phylogenetic analysis; protein coding genes; repeat sequence.
Conflict of interest statement
The authors declare no conflict of interest. The authors declare that they have no conflict of interest.
Figures






Similar articles
-
Characterization and comparative analysis of six complete mitochondrial genomes from ectomycorrhizal fungi of the Lactarius genus and phylogenetic analysis of the Agaricomycetes.Int J Biol Macromol. 2019 Jan;121:249-260. doi: 10.1016/j.ijbiomac.2018.10.029. Epub 2018 Oct 9. Int J Biol Macromol. 2019. PMID: 30308282
-
Characterization and comparative mitogenomic analysis of six newly sequenced mitochondrial genomes from ectomycorrhizal fungi (Russula) and phylogenetic analysis of the Agaricomycetes.Int J Biol Macromol. 2018 Nov;119:792-802. doi: 10.1016/j.ijbiomac.2018.07.197. Epub 2018 Aug 1. Int J Biol Macromol. 2018. PMID: 30076929
-
Analysis of the complete mitochondrial genome of Pochonia chlamydosporia suggests a close relationship to the invertebrate-pathogenic fungi in Hypocreales.BMC Microbiol. 2015 Jan 31;15:5. doi: 10.1186/s12866-015-0341-8. BMC Microbiol. 2015. PMID: 25636983 Free PMC article.
-
The complete mitochondrial genomes of five important medicinal Ganoderma species: Features, evolution, and phylogeny.Int J Biol Macromol. 2019 Oct 15;139:397-408. doi: 10.1016/j.ijbiomac.2019.08.003. Epub 2019 Aug 2. Int J Biol Macromol. 2019. PMID: 31381907
-
Hemipteran mitochondrial genomes: features, structures and implications for phylogeny.Int J Mol Sci. 2015 Jun 1;16(6):12382-404. doi: 10.3390/ijms160612382. Int J Mol Sci. 2015. PMID: 26039239 Free PMC article. Review.
Cited by
-
Mitogenomics, phylogeny and morphology reveal two new entomopathogenic species of Ophiocordyceps (Ophiocordycipitaceae, Hypocreales) from south-western China.MycoKeys. 2024 Sep 26;109:49-72. doi: 10.3897/mycokeys.109.124975. eCollection 2024. MycoKeys. 2024. PMID: 39372080 Free PMC article.
-
Morphology, phylogeny, mitogenomics and metagenomics reveal a new entomopathogenic fungus Ophiocordycepsnujiangensis (Hypocreales, Ophiocordycipitaceae) from Southwestern China.MycoKeys. 2022 Dec 21;94:91-108. doi: 10.3897/mycokeys.94.89425. eCollection 2022. MycoKeys. 2022. PMID: 36760544 Free PMC article.
-
Insights into the Deep Phylogeny and Novel Divergence Time Estimation of Patellogastropoda from Complete Mitogenomes.Genes (Basel). 2022 Jul 18;13(7):1273. doi: 10.3390/genes13071273. Genes (Basel). 2022. PMID: 35886056 Free PMC article.
-
Mitochondrial Genomes from Fungal the Entomopathogenic Moelleriella Genus Reveals Evolutionary History, Intron Dynamics and Phylogeny.J Fungi (Basel). 2025 Jan 24;11(2):94. doi: 10.3390/jof11020094. J Fungi (Basel). 2025. PMID: 39997388 Free PMC article.
-
The first two mitochondrial genomes from Apiotrichum reveal mitochondrial evolution and different taxonomic assignment of Trichosporonales.IMA Fungus. 2023 Mar 31;14(1):7. doi: 10.1186/s43008-023-00112-x. IMA Fungus. 2023. PMID: 37004131 Free PMC article.
References
-
- Rossman A.Y., Seifert K.A., Samuels G.J., Minnis A.M., Schroers H.-J., Lombard L., Crous P.W., Põldmaa K., Cannon P.F., Summerbell R.C., et al. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus. 2013;4:41–51. doi: 10.5598/imafungus.2013.04.01.05. - DOI - PMC - PubMed
-
- Domsch K.H., Gams W., Anderson T. Compendium of Soil Fungi. 2nd ed. IHW Verlag; Eching, Germany: 2007.
-
- Schroers H.J. A monograph of Bionectria (Ascomycota, Hypocreales, Bionectriaceae) and its Clonostachys anamorphs. Stud. Mycol. 2001;46:1–214.
-
- Chen W.H., Han Y.F., Liang J.D., Zou X., Liang Z.Q., Jin D.C. A new araneogenous fungus of the genus Clonostachys. Mycosystema. A new araneogenous fungus of the genus Clonostachys. Mycosystema. 2016;35:1061–1069.
-
- Zeng Z.Q., Zhuang W.Y. A new holomorphic species of Mariannaea and epitypification of M. samuelsii. Mycol. Prog. 2014;13:967–973. doi: 10.1007/s11557-014-0980-4. - DOI
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

