Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2
- PMID: 34073849
- PMCID: PMC8225085
- DOI: 10.3390/cells10061303
Isolation and Establishment of a Highly Proliferative, Cancer Stem Cell-Like, and Naturally Immortalized Triple-Negative Breast Cancer Cell Line, KAIMRC2
Abstract
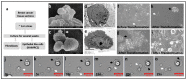
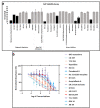
In vitro studies of a disease are key to any in vivo investigation in understanding the disease and developing new therapy regimens. Immortalized cancer cell lines are the best and easiest model for studying cancer in vitro. Here, we report the establishment of a naturally immortalized highly tumorigenic and triple-negative breast cancer cell line, KAIMRC2. This cell line is derived from a Saudi Arabian female breast cancer patient with invasive ductal carcinoma. Immunocytochemistry showed a significant ratio of the KAIMRC2 cells' expressing key breast epithelial and cancer stem cells (CSCs) markers, including CD47, CD133, CD49f, CD44, and ALDH-1A1. Gene and protein expression analysis showed overexpression of ABC transporter and AKT-PI3Kinase as well as JAK/STAT signaling pathways. In contrast, the absence of the tumor suppressor genes p53 and p73 may explain their high proliferative index. The mice model also confirmed the tumorigenic potential of the KAIMRC2 cell line, and drug tolerance studies revealed few very potent candidates. Our results confirmed an aggressive phenotype with metastatic potential and cancer stem cell-like characteristics of the KAIMR2 cell line. Furthermore, we have also presented potent small molecule inhibitors, especially Ryuvidine, that can be further developed, alone or in synergy with other potent inhibitors, to target multiple cancer-related pathways.
Keywords: KAIMRC2; breast cancer; cell line; characterization; drug treatment; metastatic; stem cells; triple negative.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Up-modulation of PLC-β2 reduces the number and malignancy of triple-negative breast tumor cells with a CD133+/EpCAM+ phenotype: a promising target for preventing progression of TNBC.BMC Cancer. 2017 Sep 4;17(1):617. doi: 10.1186/s12885-017-3592-y. BMC Cancer. 2017. PMID: 28870198 Free PMC article.
-
The Milk Protein Alpha-Casein Suppresses Triple Negative Breast Cancer Stem Cell Activity Via STAT and HIF-1alpha Signalling Pathways in Breast Cancer Cells and Fibroblasts.J Mammary Gland Biol Neoplasia. 2019 Sep;24(3):245-256. doi: 10.1007/s10911-019-09435-1. Epub 2019 Sep 12. J Mammary Gland Biol Neoplasia. 2019. PMID: 31529195
-
EP300 knockdown reduces cancer stem cell phenotype, tumor growth and metastasis in triple negative breast cancer.BMC Cancer. 2020 Nov 10;20(1):1076. doi: 10.1186/s12885-020-07573-y. BMC Cancer. 2020. PMID: 33167919 Free PMC article.
-
Identification of a stemness-related gene panel associated with BET inhibition in triple negative breast cancer.Cell Oncol (Dordr). 2020 Jun;43(3):431-444. doi: 10.1007/s13402-020-00497-6. Epub 2020 Mar 12. Cell Oncol (Dordr). 2020. PMID: 32166583 Free PMC article.
-
Signaling pathways essential for triple-negative breast cancer stem-like cells.Stem Cells. 2021 Feb;39(2):133-143. doi: 10.1002/stem.3301. Epub 2020 Dec 8. Stem Cells. 2021. PMID: 33211379 Review.
Cited by
-
Bioactive silver nanoparticles fabricated using Lasiurus scindicus and Panicum turgidum seed extracts: anticancer and antibacterial efficiency.Sci Rep. 2024 Feb 20;14(1):4162. doi: 10.1038/s41598-024-54449-3. Sci Rep. 2024. PMID: 38378923 Free PMC article.
-
Pharmacological Profiling of Calotropis Procera and Rhazya Stricta: Unraveling the Antibacterial and Anti-Cancer Potential of Chemically Active Metabolites.J Cancer. 2025 Jan 1;16(1):12-33. doi: 10.7150/jca.96848. eCollection 2025. J Cancer. 2025. PMID: 39744559 Free PMC article.
-
Discovery of a novel potentially transforming somatic mutation in CSF2RB gene in breast cancer.Cancer Med. 2021 Nov;10(22):8138-8150. doi: 10.1002/cam4.4106. Epub 2021 Nov 2. Cancer Med. 2021. PMID: 34729943 Free PMC article.
-
Bestatin attenuates breast cancer stemness by targeting puromycin-sensitive aminopeptidase.Discov Oncol. 2024 May 30;15(1):197. doi: 10.1007/s12672-024-01063-4. Discov Oncol. 2024. PMID: 38814491 Free PMC article.
-
Aspergillus Species from the Sabkha Marsh: Potential Antimicrobial and Anticancer Agents Revealed Through Molecular and Pharmacological Analysis.Biologics. 2024 Aug 6;18:207-228. doi: 10.2147/BTT.S472491. eCollection 2024. Biologics. 2024. PMID: 39130166 Free PMC article.
References
-
- American Cancer Society . Global Cancer Facts & Figures. 4th ed. American Cancer Society; Atlanta, GA, USA: 2018. pp. 1–76.
-
- Chopra S., Davies E.L. Breast cancer. Medicine. 2020;48:113–118. doi: 10.1016/j.mpmed.2019.11.009. - DOI
-
- Saudi Cancer Registry . Saudi Cancer Registry Cancer Incidence Report Saudi Arabia, 2014. Saudi Health Council; Riyadh, Saudi Arabia: 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

