Ancient Bacterial Class Alphaproteobacteria Cytochrome P450 Monooxygenases Can Be Found in Other Bacterial Species
- PMID: 34073951
- PMCID: PMC8197338
- DOI: 10.3390/ijms22115542
Ancient Bacterial Class Alphaproteobacteria Cytochrome P450 Monooxygenases Can Be Found in Other Bacterial Species
Abstract
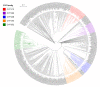
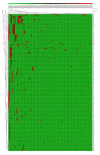
Cytochrome P450 monooxygenases (CYPs/P450s), heme-thiolate proteins, are well-known players in the generation of chemicals valuable to humans and as a drug target against pathogens. Understanding the evolution of P450s in a bacterial population is gaining momentum. In this study, we report comprehensive analysis of P450s in the ancient group of the bacterial class Alphaproteobacteria. Genome data mining and annotation of P450s in 599 alphaproteobacterial species belonging to 164 genera revealed the presence of P450s in only 241 species belonging to 82 genera that are grouped into 143 P450 families and 214 P450 subfamilies, including 77 new P450 families. Alphaproteobacterial species have the highest average number of P450s compared to Firmicutes species and cyanobacterial species. The lowest percentage of alphaproteobacterial species P450s (2.4%) was found to be part of secondary metabolite biosynthetic gene clusters (BGCs), compared other bacterial species, indicating that during evolution large numbers of P450s became part of BGCs in other bacterial species. Our study identified that some of the P450 families found in alphaproteobacterial species were passed to other bacterial species. This is the first study to report on the identification of CYP125 P450, cholesterol and cholest-4-en-3-one hydroxylase in alphaproteobacterial species (Phenylobacterium zucineum) and to predict cholesterol side-chain oxidation capability (based on homolog proteins) by P. zucineum.
Keywords: Alphaproteobacteria; CYP125; annotation; biosynthetic gene clusters; cholesterol oxidation; cytochrome P450 monooxygenase; genome data mining.
Conflict of interest statement
The authors declare no conflict of interest and the funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures



References
-
- Yamazaki H. Fifty Years of Cytochrome P450 Research. Springer Japan; Tokyo, Japan: 2014. pp. 1–409.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

