High Concordance of Genomic Profiles between Primary and Metastatic Colorectal Cancer
- PMID: 34074070
- PMCID: PMC8197329
- DOI: 10.3390/ijms22115561
High Concordance of Genomic Profiles between Primary and Metastatic Colorectal Cancer
Abstract
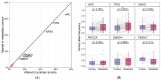
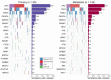
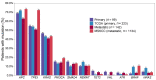
The comparison of the genetic profiles between primary and metastatic colorectal cancer (CRC) is needed to enable the discovery of useful therapeutic targets against metastatic CRCs. We performed the targeted next generation sequencing assay of 170 cancer-associated genes for 142 metastatic CRCs, including 95 pairs of primary and metastatic CRCs, to reveal their genomic characteristics and to assess the genetic heterogeneity. The most frequently mutated gene in primary and metastatic CRCs was APC (71% vs. 65%), TP53 (54% vs. 57%), KRAS (45% vs. 44%), PIK3CA (16% vs. 19%), SMAD4 (15% vs. 14%) and FBXW7 (11% vs. 11%). The concordance in the top six frequently mutated genes was 85%, on average. The overall mutation frequencies were consistent with two sets of public data (TCGA and MSKCC). To the author's knowledge, this is the first study to compare the genetic profiles of our cohort with that of the metastatic CRCs from MSKCC. Comparative sequencing analysis between primary and metastatic CRCs revealed a high degree of genetic concordance in the current clinically actionable genes. Therefore, the genetic investigation of archived primary tumor samples with the challenges of obtaining an adequate sample from metastatic sites appears to be sufficient for the application of cancer precision medicine in the metastatic setting.
Keywords: genomic profiles; high concordance between primary and metastatic colorectal cancer; metastatic colorectal cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Yaeger R., Chatila W.K., Lipsyc M.D., Hechtman J.F., Cercek A., Sanchez-Vega F., Jayakumaran G., Middha S., Zehir A., Donoghue M.T.A., et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33:125–136 e3. doi: 10.1016/j.ccell.2017.12.004. - DOI - PMC - PubMed
-
- Jones S., Chen W.D., Parmigiani G., Diehl F., Beerenwinkel N., Antal T., Traulsen A., Nowak M.A., Siegel C., Velculescu V.E., et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

