Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters
- PMID: 34074255
- PMCID: PMC8169432
- DOI: 10.1186/s12920-021-00990-3
Molecular epidemiology of SARS-CoV-2 isolated from COVID-19 family clusters
Abstract
Background: Transmission within families and multiple spike protein mutations have been associated with the rapid transmission of SARS-CoV-2. We aimed to: (1) describe full genome characterization of SARS-CoV-2 and correlate the sequences with epidemiological data within family clusters, and (2) conduct phylogenetic analysis of all samples from Yogyakarta and Central Java, Indonesia and other countries.
Methods: The study involved 17 patients with COVID-19, including two family clusters. We determined the full-genome sequences of SARS-CoV-2 using the Illumina MiSeq next-generation sequencer. Phylogenetic analysis was performed using a dataset of 142 full-genomes of SARS-CoV-2 from different regions.
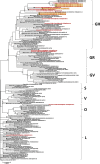
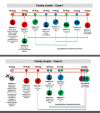
Results: Ninety-four SNPs were detected throughout the open reading frame (ORF) of SARS-CoV-2 samples with 58% (54/94) of the nucleic acid changes resulting in amino acid mutations. About 94% (16/17) of the virus samples showed D614G on spike protein and 56% of these (9/16) showed other various amino acid mutations on this protein, including L5F, V83L, V213A, W258R, Q677H, and N811I. The virus samples from family cluster-1 (n = 3) belong to the same clade GH, in which two were collected from deceased patients, and the other from the survived patient. All samples from this family cluster revealed a combination of spike protein mutations of D614G and V213A. Virus samples from family cluster-2 (n = 3) also belonged to the clade GH and showed other spike protein mutations of L5F alongside the D614G mutation.
Conclusions: Our study is the first comprehensive report associating the full-genome sequences of SARS-CoV-2 with the epidemiological data within family clusters. Phylogenetic analysis revealed that the three viruses from family cluster-1 formed a monophyletic group, whereas viruses from family cluster-2 formed a polyphyletic group indicating there is the possibility of different sources of infection. This study highlights how the same spike protein mutations among members of the same family might show different disease outcomes.
Keywords: COVID-19 severity; Family cluster; Multiple spike protein mutations; Phylogenetic analysis; SARS-CoV-2 transmission; Whole genome sequencing.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

References
-
- World Health Organization. https://www.who.int/news-room/detail/27-04-2020-who-timeline---covid-19. Accessed 9 Feb 2021.
-
- World Health Organization. https://covid19.who.int/table. Accessed 9 Feb 2021.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

