The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway
- PMID: 34074294
- PMCID: PMC8167978
- DOI: 10.1186/s12943-021-01375-x
The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway
Abstract
Background: Noncoding RNAs such as circular RNAs (circRNAs) are abundant in the human body and influence the occurrence and development of various diseases. However, the biological functions of circRNAs in colorectal cancer (CRC) are largely unknown.
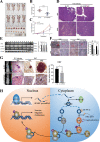
Methods: RT-qPCR was used to detect the expression of circRNAs and mRNA in CRC cells and tissues. Fluorescence in situ hybridization (FISH) was used to analyze the location of circSPARC. Function-based experiments were performed using circSPARC knockdown and overexpression cell lines in vitro and in vivo, including CCK8, colony formation, transwell and metastasis models. Mechanistically, luciferase reporter assay, western blots, RNA immunoprecipitation (RIP), Chromatin isolation by RNA purification (ChIRP) and immunohistochemical stainings were performed.
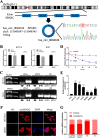
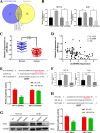
Results: CircSPARC was upregulated in both the tissues and plasma of CRC patients. High expression of circSPARC was associated with advanced TNM stage, lymph node metastases, and poor survival. Silencing circSPARC inhibited CRC cell migration and proliferation in vitro and vivo. Mechanistically, circSPARC sponged miR-485-3p to upregulate JAK2 expression and ultimately contribute to the accumulation of phosphorylated (p)-STAT3. Besides, circSPARC recruited FUS, which facilitated the nuclear translocation of p-STAT3.
Conclusions: These findings suggest that circSPARC might serve as a potential diagnostic and prognostic biomarker and a therapeutic target for CRC treatment by regulating JAK2/STAT3 pathway.
Keywords: Biomarker; Colorectal cancer; JAK/STAT signalling pathway; circRNA; circSPARC.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures





Similar articles
-
Circular RNA circ_0000372 contributes to the proliferation, migration and invasion of colorectal cancer by elevating IL6 expression via sponging miR-495.Anticancer Drugs. 2021 Mar 1;32(3):296-305. doi: 10.1097/CAD.0000000000001002. Anticancer Drugs. 2021. PMID: 33534412
-
The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation.Mol Cancer. 2020 Nov 23;19(1):164. doi: 10.1186/s12943-020-01272-9. Mol Cancer. 2020. PMID: 33225938 Free PMC article.
-
EIF4A3-induced circ_0084615 contributes to the progression of colorectal cancer via miR-599/ONECUT2 pathway.J Exp Clin Cancer Res. 2021 Jul 12;40(1):227. doi: 10.1186/s13046-021-02029-y. J Exp Clin Cancer Res. 2021. PMID: 34253241 Free PMC article.
-
Crosstalk between circular RNAs and the STAT3 signaling pathway in human cancer.Biochim Biophys Acta Gene Regul Mech. 2024 Dec;1867(4):195051. doi: 10.1016/j.bbagrm.2024.195051. Epub 2024 Aug 8. Biochim Biophys Acta Gene Regul Mech. 2024. PMID: 39121909 Review.
-
Circular RNA in colorectal cancer.J Cell Mol Med. 2021 Apr;25(8):3667-3679. doi: 10.1111/jcmm.16380. Epub 2021 Mar 9. J Cell Mol Med. 2021. PMID: 33687140 Free PMC article. Review.
Cited by
-
Deciphering treatment resistance in metastatic colorectal cancer: roles of drug transports, EGFR mutations, and HGF/c-MET signaling.Front Pharmacol. 2024 Jan 10;14:1340401. doi: 10.3389/fphar.2023.1340401. eCollection 2023. Front Pharmacol. 2024. PMID: 38269272 Free PMC article. Review.
-
The Role of JAK-STAT-SOCS1 Axis in Tumorigenesis, Malignant Progression and Lymphatic Metastasis of Penile Cancer.Int J Med Sci. 2024 Apr 29;21(6):1176-1186. doi: 10.7150/ijms.95490. eCollection 2024. Int J Med Sci. 2024. PMID: 38774752 Free PMC article.
-
Mechanisms and therapeutic implications of gene expression regulation by circRNA-protein interactions in cancer.Commun Biol. 2025 Jan 17;8(1):77. doi: 10.1038/s42003-024-07383-z. Commun Biol. 2025. PMID: 39825074 Free PMC article. Review.
-
circCAPRIN1 interacts with STAT2 to promote tumor progression and lipid synthesis via upregulating ACC1 expression in colorectal cancer.Cancer Commun (Lond). 2023 Jan;43(1):100-122. doi: 10.1002/cac2.12380. Epub 2022 Nov 3. Cancer Commun (Lond). 2023. PMID: 36328987 Free PMC article.
-
Hypoxia-Induced FUS-circTBC1D14 Stress Granules Promote Autophagy in TNBC.Adv Sci (Weinh). 2023 Apr;10(10):e2204988. doi: 10.1002/advs.202204988. Epub 2023 Feb 19. Adv Sci (Weinh). 2023. PMID: 36806670 Free PMC article.
References
-
- Brody H. Colorectal cancer. Nature. 521(2015):S1. - PubMed
-
- Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, van Groeningen CJ, Sinnige HA, Richel DJ, Voest EE, Dijkstra JR, Vink-Börger ME, Antonini NF, Mol L, van Krieken JH, Dalesio O, Punt CJ. Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med. 2009;360(6):563–572. doi: 10.1056/NEJMoa0808268. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

