The energy-spectrum of bicompatible sequences
- PMID: 34074304
- PMCID: PMC8167974
- DOI: 10.1186/s13015-021-00187-4
The energy-spectrum of bicompatible sequences
Abstract
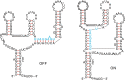
Background: Genotype-phenotype maps provide a meaningful filtration of sequence space and RNA secondary structures are particular such phenotypes. Compatible sequences, which satisfy the base-pairing constraints of a given RNA structure, play an important role in the context of neutral evolution. Sequences that are simultaneously compatible with two given structures (bicompatible sequences), are beacons in phenotypic transitions, induced by erroneously replicating populations of RNA sequences. RNA riboswitches, which are capable of expressing two distinct secondary structures without changing the underlying sequence, are one example of bicompatible sequences in living organisms.
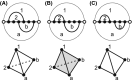
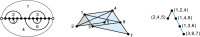
Results: We present a full loop energy model Boltzmann sampler of bicompatible sequences for pairs of structures. The sequence sampler employs a dynamic programming routine whose time complexity is polynomial when assuming the maximum number of exposed vertices, [Formula: see text], is a constant. The parameter [Formula: see text] depends on the two structures and can be very large. We introduce a novel topological framework encapsulating the relations between loops that sheds light on the understanding of [Formula: see text]. Based on this framework, we give an algorithm to sample sequences with minimum [Formula: see text] on a particular topologically classified case as well as giving hints to the solution in the other cases. As a result, we utilize our sequence sampler to study some established riboswitches.
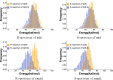
Conclusion: Our analysis of riboswitch sequences shows that a pair of structures needs to satisfy key properties in order to facilitate phenotypic transitions and that pairs of random structures are unlikely to do so. Our analysis observes a distinct signature of riboswitch sequences, suggesting a new criterion for identifying native sequences and sequences subjected to evolutionary pressure. Our free software is available at: https://github.com/FenixHuang667/Bifold .
Keywords: Evolutionary transition; Riboswitch; Topological nerve.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures









Similar articles
-
An Efficient Dual Sampling Algorithm with Hamming Distance Filtration.J Comput Biol. 2018 Nov;25(11):1179-1192. doi: 10.1089/cmb.2018.0075. Epub 2018 Aug 22. J Comput Biol. 2018. PMID: 30133328
-
A Boltzmann Sampler for 1-Pairs with Double Filtration.J Comput Biol. 2019 Mar;26(3):173-192. doi: 10.1089/cmb.2018.0095. Epub 2019 Jan 17. J Comput Biol. 2019. PMID: 30653353
-
An improved Four-Russians method and sparsified Four-Russians algorithm for RNA folding.Algorithms Mol Biol. 2016 Aug 5;11:22. doi: 10.1186/s13015-016-0081-9. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27499801 Free PMC article.
-
Sparse RNA folding revisited: space-efficient minimum free energy structure prediction.Algorithms Mol Biol. 2016 Apr 23;11:7. doi: 10.1186/s13015-016-0071-y. eCollection 2016. Algorithms Mol Biol. 2016. PMID: 27110275 Free PMC article. Review.
-
How to search for RNA structures. Theoretical concepts in evolutionary biotechnology.J Biotechnol. 1995 Jul 31;41(2-3):239-57. doi: 10.1016/0168-1656(94)00085-q. J Biotechnol. 1995. PMID: 7544591 Review.
References
-
- Grüner W, Giegerich R, Strothmann D, Reidys C, Weber J, Hofacker IL, Stadler PF, Schuster P. Analysis of RNA sequence structure maps by exhaustive enumeration I. Neutral networks. Monatsh Chem. 1996;127:355–374. doi: 10.1007/BF00810881. - DOI
-
- Grüner W, Giegerich R, Strothmann D, Reidys C, Weber J, Hofacker IL, Stadler PF, Schuster P. Analysis of RNA sequence structure maps by exhaustive enumeration II. Structures of neutral networks and shape space covering. Monatsh Chem. 1996;127:375–389. doi: 10.1007/BF00810882. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

