Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids
- PMID: 34074327
- PMCID: PMC8167833
- DOI: 10.1186/s13073-021-00912-z
Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids
Abstract
Background: Metagenomic next-generation sequencing (mNGS) of body fluids is an emerging approach to identify occult pathogens in undiagnosed patients. We hypothesized that metagenomic testing can be simultaneously used to detect malignant neoplasms in addition to infectious pathogens.
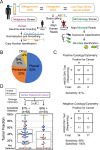
Methods: From two independent studies (n = 205), we used human data generated from a metagenomic sequencing pipeline to simultaneously screen for malignancies by copy number variation (CNV) detection. In the first case-control study, we analyzed body fluid samples (n = 124) from patients with a clinical diagnosis of either malignancy (positive cases, n = 65) or infection (negative controls, n = 59). In a second verification cohort, we analyzed a series of consecutive cases (n = 81) sent to cytology for malignancy workup that included malignant positives (n = 32), negatives (n = 18), or cases with an unclear gold standard (n = 31).
Results: The overall CNV test sensitivity across all studies was 87% (55 of 63) in patients with malignancies confirmed by conventional cytology and/or flow cytometry testing and 68% (23 of 34) in patients who were ultimately diagnosed with cancer but negative by conventional testing. Specificity was 100% (95% CI 95-100%) with no false positives detected in 77 negative controls. In one example, a patient hospitalized with an unknown pulmonary illness had non-diagnostic lung biopsies, while CNVs implicating a malignancy were detectable from bronchoalveolar fluid.
Conclusions: Metagenomic sequencing of body fluids can be used to identify undetected malignant neoplasms through copy number variation detection. This study illustrates the potential clinical utility of a single metagenomic test to uncover the cause of undiagnosed acute illnesses due to cancer or infection using the same specimen.
Conflict of interest statement
ET was employed by DNAnexus, Inc. during the duration of the study and by Karius, Inc. prior to publication. The remaining authors declare that they have no competing interests.
Figures

References
-
- Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu G, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, Berger A, Douglas V, Josephson SA, Chow FC, Fulton BD, DeRisi JL, Gelfand JM, Naccache SN, Bender J, Dien Bard J, Murkey J, Carlson M, Vespa PM, Vijayan T, Allyn PR, Campeau S, Humphries RM, Klausner JD, Ganzon CD, Memar F, Ocampo NA, Zimmermann LL, Cohen SH, Polage CR, DeBiasi RL, Haller B, Dallas R, Maron G, Hayden R, Messacar K, Dominguez SR, Miller S, Chiu CY. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med. 2019;380(24):2327–2340. doi: 10.1056/NEJMoa1803396. - DOI - PMC - PubMed
-
- Miller S, Naccache SN, Samayoa E, Messacar K, Arevalo S, Federman S, et al. Laboratory validation of a clinical metagenomic sequencing assay for pathogen detection in cerebrospinal fluid. Genome Res. 2019; Available from: http://genome.cshlp.org/content/early/2019/03/07/gr.238170.118. [cited 2019 May 31]. - PMC - PubMed
-
- Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD, Cheung A, Rogers ZN, Meshulam-Simon G, Huijse L, Balakrishnan S, Quinn JV, Hollemon D, Hong DK, Vaughn ML, Kertesz M, Bercovici S, Wilber JC, Yang S. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. doi: 10.1038/s41564-018-0349-6. - DOI - PubMed
-
- Goggin KP, Gonzalez-Pena V, Inaba Y, Allison KJ, Hong DK, Ahmed AA, et al. Evaluation of plasma microbial cell-free DNA sequencing to predict bloodstream infection in pediatric patients with relapsed or refractory cancer. JAMA Oncol. 2019; Available from: https://jamanetwork.com/journals/jamaoncology/fullarticle/2757390. [cited 2020 Jan 4] - PMC - PubMed

