Generation of ordered protein assemblies using rigid three-body fusion
- PMID: 34074752
- PMCID: PMC8201882
- DOI: 10.1073/pnas.2015037118
Generation of ordered protein assemblies using rigid three-body fusion
Abstract
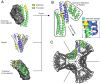
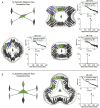
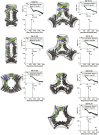
Protein nanomaterial design is an emerging discipline with applications in medicine and beyond. A long-standing design approach uses genetic fusion to join protein homo-oligomer subunits via α-helical linkers to form more complex symmetric assemblies, but this method is hampered by linker flexibility and a dearth of geometric solutions. Here, we describe a general computational method for rigidly fusing homo-oligomer and spacer building blocks to generate user-defined architectures that generates far more geometric solutions than previous approaches. The fusion junctions are then optimized using Rosetta to minimize flexibility. We apply this method to design and test 92 dihedral symmetric protein assemblies using a set of designed homodimers and repeat protein building blocks. Experimental validation by native mass spectrometry, small-angle X-ray scattering, and negative-stain single-particle electron microscopy confirms the assembly states for 11 designs. Most of these assemblies are constructed from designed ankyrin repeat proteins (DARPins), held in place on one end by α-helical fusion and on the other by a designed homodimer interface, and we explored their use for cryogenic electron microscopy (cryo-EM) structure determination by incorporating DARPin variants selected to bind targets of interest. Although the target resolution was limited by preferred orientation effects and small scaffold size, we found that the dual anchoring strategy reduced the flexibility of the target-DARPIN complex with respect to the overall assembly, suggesting that multipoint anchoring of binding domains could contribute to cryo-EM structure determination of small proteins.
Keywords: DARPin; cryo-EM; nanomaterials; protein design; protein fusion.
Conflict of interest statement
The authors declare no competing interest.
Figures

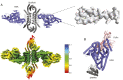
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

