Solving patients with rare diseases through programmatic reanalysis of genome-phenome data
- PMID: 34075210
- PMCID: PMC8440686
- DOI: 10.1038/s41431-021-00852-7
Solving patients with rare diseases through programmatic reanalysis of genome-phenome data
Erratum in
-
Correction to: Solving patients with rare diseases through programmatic reanalysis of genome-phenome data.Eur J Hum Genet. 2021 Sep;29(9):1466-1469. doi: 10.1038/s41431-021-00934-6. Eur J Hum Genet. 2021. PMID: 34393220 Free PMC article. No abstract available.
Abstract
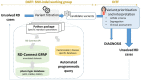
Reanalysis of inconclusive exome/genome sequencing data increases the diagnosis yield of patients with rare diseases. However, the cost and efforts required for reanalysis prevent its routine implementation in research and clinical environments. The Solve-RD project aims to reveal the molecular causes underlying undiagnosed rare diseases. One of the goals is to implement innovative approaches to reanalyse the exomes and genomes from thousands of well-studied undiagnosed cases. The raw genomic data is submitted to Solve-RD through the RD-Connect Genome-Phenome Analysis Platform (GPAP) together with standardised phenotypic and pedigree data. We have developed a programmatic workflow to reanalyse genome-phenome data. It uses the RD-Connect GPAP's Application Programming Interface (API) and relies on the big-data technologies upon which the system is built. We have applied the workflow to prioritise rare known pathogenic variants from 4411 undiagnosed cases. The queries returned an average of 1.45 variants per case, which first were evaluated in bulk by a panel of disease experts and afterwards specifically by the submitter of each case. A total of 120 index cases (21.2% of prioritised cases, 2.7% of all exome/genome-negative samples) have already been solved, with others being under investigation. The implementation of solutions as the one described here provide the technical framework to enable periodic case-level data re-evaluation in clinical settings, as recommended by the American College of Medical Genetics.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

