Structural insights and inhibition mechanism of TMPRSS2 by experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to control SARS-coronavirus-2: A molecular modeling approach
- PMID: 34075338
- PMCID: PMC8152215
- DOI: 10.1016/j.imu.2021.100597
Structural insights and inhibition mechanism of TMPRSS2 by experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to control SARS-coronavirus-2: A molecular modeling approach
Abstract
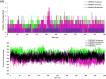
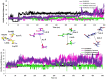
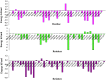
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) has been responsible for the cause of global pandemic Covid-19 and to date, there is no effective treatment available. The spike 'S' protein of SARS-CoV-2 and ACE2 of the host cell are being targeted to design new drugs to control Covid-19. Similarly, a transmembrane serine protease, TMPRSS2 of the host cell plays a significant role in the proteolytic cleavage of viral 'S' protein helpful for the priming of ACE2 receptors and viral entry into human cells. However, three-dimensional structural information and the inhibition mechanism of TMPRSS2 is yet to be explored experimentally. Hence, we have used a molecular dynamics (MD) simulated homology model of TMPRSS2 to study the inhibition mechanism of experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride (BHH) using molecular modeling techniques. Prior to docking, all three inhibitors were geometry optimized by semi-empirical quantum chemical RM1 method. Molecular docking analysis revealed that Camostat mesylate and its structural analogue Nafamostat interact strongly with residues His296 and Ser441 present in the catalytic triad of TMPRSS2, whereas BHH binds with Ala386 along with other residues. Comparative molecular dynamics simulations revealed the stable behavior of all the docked complexes. MM-PBSA calculations also revealed the stronger binding of Camostat mesylate to TMPRSS2 active site residues as compared to Nafamostat and BHH. Thus, this structural information could be useful to understand the mechanistic approach of TMPRSS2 inhibition, which may be helpful to design new lead compounds to prevent the entry of SARS-Coronavirus 2 in human cells.
Keywords: Covid-19; Molecular docking; Molecular dynamics simulation; SARS-CoV-2; TMPRSS2.
© 2021 Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures









References
-
- WHO WHO Director-General opening remarks at the media briefing on COVID-19-11 March 2020. 2020. https://www.who.int/director-general/speeches/detail/who-director-genera... [Online] Availabble.
LinkOut - more resources
Full Text Sources
Miscellaneous
