Mantis: flexible and consensus-driven genome annotation
- PMID: 34076241
- PMCID: PMC8170692
- DOI: 10.1093/gigascience/giab042
Mantis: flexible and consensus-driven genome annotation
Abstract
Background: The rapid development of the (meta-)omics fields has produced an unprecedented amount of high-resolution and high-fidelity data. Through the use of these datasets we can infer the role of previously functionally unannotated proteins from single organisms and consortia. In this context, protein function annotation can be described as the identification of regions of interest (i.e., domains) in protein sequences and the assignment of biological functions. Despite the existence of numerous tools, challenges remain in terms of speed, flexibility, and reproducibility. In the big data era, it is also increasingly important to cease limiting our findings to a single reference, coalescing knowledge from different data sources, and thus overcoming some limitations in overly relying on computationally generated data from single sources.
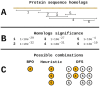
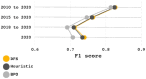
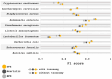
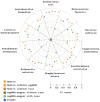
Results: We implemented a protein annotation tool, Mantis, which uses database identifiers intersection and text mining to integrate knowledge from multiple reference data sources into a single consensus-driven output. Mantis is flexible, allowing for the customization of reference data and execution parameters, and is reproducible across different research goals and user environments. We implemented a depth-first search algorithm for domain-specific annotation, which significantly improved annotation performance compared to sequence-wide annotation. The parallelized implementation of Mantis results in short runtimes while also outputting high coverage and high-quality protein function annotations.
Conclusions: Mantis is a protein function annotation tool that produces high-quality consensus-driven protein annotations. It is easy to set up, customize, and use, scaling from single genomes to large metagenomes. Mantis is available under the MIT license at https://github.com/PedroMTQ/mantis.
Keywords: HMM; bioinformatics; consensus; homology; protein function annotation.
© The Author(s) 2021. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
COGNIZER: A Framework for Functional Annotation of Metagenomic Datasets.PLoS One. 2015 Nov 11;10(11):e0142102. doi: 10.1371/journal.pone.0142102. eCollection 2015. PLoS One. 2015. PMID: 26561344 Free PMC article.
-
Comprehensive Functional Annotation of Metagenomes and Microbial Genomes Using a Deep Learning-Based Method.mSystems. 2023 Apr 27;8(2):e0117822. doi: 10.1128/msystems.01178-22. Epub 2023 Mar 7. mSystems. 2023. PMID: 37010293 Free PMC article.
-
MicrobeAnnotator: a user-friendly, comprehensive functional annotation pipeline for microbial genomes.BMC Bioinformatics. 2021 Jan 6;22(1):11. doi: 10.1186/s12859-020-03940-5. BMC Bioinformatics. 2021. PMID: 33407081 Free PMC article.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
Folic acid supplementation and malaria susceptibility and severity among people taking antifolate antimalarial drugs in endemic areas.Cochrane Database Syst Rev. 2022 Feb 1;2(2022):CD014217. doi: 10.1002/14651858.CD014217. Cochrane Database Syst Rev. 2022. PMID: 36321557 Free PMC article.
Cited by
-
Challenges, Strategies, and Perspectives for Reference-Independent Longitudinal Multi-Omic Microbiome Studies.Front Genet. 2021 Jun 14;12:666244. doi: 10.3389/fgene.2021.666244. eCollection 2021. Front Genet. 2021. PMID: 34194470 Free PMC article.
-
Functional prediction of proteins from the human gut archaeome.ISME Commun. 2024 Jan 10;4(1):ycad014. doi: 10.1093/ismeco/ycad014. eCollection 2024 Jan. ISME Commun. 2024. PMID: 38486809 Free PMC article.
-
Microbial communities reveal niche partitioning across the slope and bottom zones of the challenger deep.Environ Microbiol Rep. 2024 Aug;16(4):e13314. doi: 10.1111/1758-2229.13314. Environ Microbiol Rep. 2024. PMID: 39086173 Free PMC article.
-
Phylogenomic Analyses of Snodgrassella Isolates from Honeybees and Bumblebees Reveal Taxonomic and Functional Diversity.mSystems. 2022 Jun 28;7(3):e0150021. doi: 10.1128/msystems.01500-21. Epub 2022 May 23. mSystems. 2022. PMID: 35604118 Free PMC article.
-
Critical Assessment of MetaProteome Investigation (CAMPI): a multi-laboratory comparison of established workflows.Nat Commun. 2021 Dec 15;12(1):7305. doi: 10.1038/s41467-021-27542-8. Nat Commun. 2021. PMID: 34911965 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

