Recent Advances in the Development of Sigma Receptor Ligands as Cytotoxic Agents: A Medicinal Chemistry Perspective
- PMID: 34076441
- PMCID: PMC8279423
- DOI: 10.1021/acs.jmedchem.0c02265
Recent Advances in the Development of Sigma Receptor Ligands as Cytotoxic Agents: A Medicinal Chemistry Perspective
Abstract
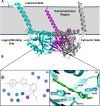
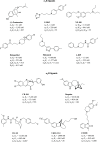
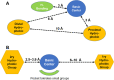
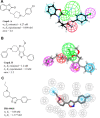
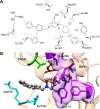
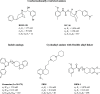
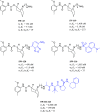
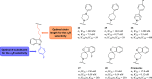
Since their discovery as distinct receptor proteins, the specific physiopathological role of sigma receptors (σRs) has been deeply investigated. It has been reported that these proteins, classified into two subtypes indicated as σ1 and σ2, might play a pivotal role in cancer growth, cell proliferation, and tumor aggressiveness. As a result, the development of selective σR ligands with potential antitumor properties attracted significant attention as an emerging theme in cancer research. This perspective deals with the recent advances of σR ligands as novel cytotoxic agents, covering articles published between 2010 and 2020. An up-to-date description of the medicinal chemistry of selective σ1R and σ2R ligands with antiproliferative and cytotoxic activities has been provided, including major pharmacophore models and comprehensive structure-activity relationships for each main class of σR ligands.
Conflict of interest statement
The authors declare no competing financial interest.
Figures










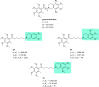
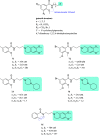
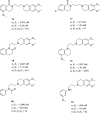
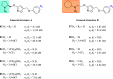
References
-
- Heron M.; Anderson R. N. Changes in the Leading Cause of Death: Recent Patterns in Heart Disease and Cancer Mortality. NCHS data Brief. 2016, 1–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

